Next Lesson - Structure of Proteins
Abstract
- DNA is packaged to fit inside the nucleus of a cell by being wrapped around histone proteins which are then further coiled into solenoids.
- Chromatin can either be condensed heterochromatin or uncondensed euchromatin.
- DNA is a polymer of nucleotides. Nucleotides are made up of a nitrogenous base, pentose sugar and phosphate group.
- The nucleotides are covalently linked together in a chain through 3’ to 5’ phosphodiester bonds.
- The nitrogenous bases can be classed as a purine or a pyrimidine and the bonding between these bases helps form the double helix structure of DNA molecule.
- DNA replication takes place in the S phase of the cell cycle and involves three stages which are initiation, elongation and termination.
Core
Deoxyribonucleic acid or DNA is a molecule, which is found in the nucleus of every cell in the human body. It has a double helix structure and it carries the genetic instructions for cell reproduction, growth, development and function.
The width of the DNA chain is 2nm, but the total length of DNA in each cell is around 2m (no you didn’t read that wrong, 2 meters in every cell!) therefore DNA needs to packaged to fit inside each cell. This is achieved by DNA being wrapped twice around a number histone proteins. A histone protein with DNA wrapped around it is called a nucleosome. Nucleosomes are packaged into a thread which under a transmission electron microscope has the classical appearance of ‘beads on a string’. The nucleosomes are further organised into higher order chromatin structures, classically described as a solenoid with a width of around 30 nm. Solenoids can be looped and coiled into chromosomes that are seen during mitosis and meiosis.
Chromatin is a complex of DNA and histones with the basic structural unit being a nucleosome. In the nucleus of the cell, there is heterochromatin and euchromatin:
- Heterochromatin is highly condensed as it is mostly made up of solenoids. As a result genes are not easily expressed in this form and as a result little RNA synthesis happens in these sections.
- Euchromatin has DNA packaged in a ‘beads on a string’ structure. As a result the genes are easily expressed and as a result more RNA synthesis happens in these sections.
Under an electron microscope, heterochromatin appears dark due to its condensed nature and euchromatin is much lighter or not visible.
A gene is a region of DNA that codes for a specific protein. Each gene has a specific location on a specific chromosome known as a locus. There are around 25,000 genes in humans spread across 23 pairs of chromosomes - 22 autosomes and a pair of sex chromosomes.
Both DNA and Ribonucleic Acid (RNA) are nucleic acids (polymer of nucleotides(be mindful of spelling)).
- NucleoSide = base + sugar
- NucleoTide = base + sugar + phosphate
The pentose sugar in DNA is 2-deoxyribose whereas the pentose sugar in RNA is ribose.
As you can see in the diagram, 2-deoxyribose has only a hydrogen atom on the second carbon (therefore no extra oxygen hence deoxyribose), whereas ribose has a hydroxyl group on second carbon instead.
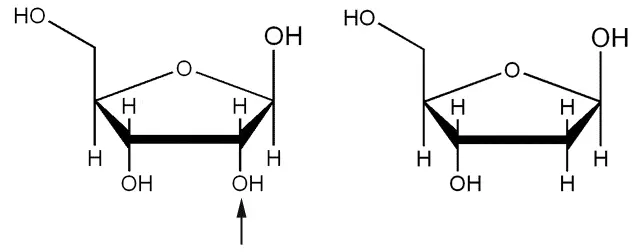
Diagram - The difference in structure between Ribose (pictured left) and Deoxyribose (pictured right)
Creative commons source by Pixie [CC BY 3.0 (https://creativecommons.org/licenses/by/3.0)]
The bases in DNA and RNA are known as nitrogenous bases and can be classed as either a purine or a pyrimidine. Purines are double-ring structures while pyrimidines are single ring structures. The purines are the same in both DNA and RNA, however, the pyrimidines used in DNA and RNA are different:
- The purines are Adenine and Guanine.
- The pyrimidines are Cytosine, Thymine and Uracil
- N.B. In DNA Thymine is used in pairs with Adenine, whereas in RNA Uracil pairs with Adenine. (for pairing rules see below)
DNA’s primary structure is that of a polymer of nucleotides covalently linked by phosphodiester bonds. These covalent bonds form the supporting ‘phosphate backbone’. The secondary structure of DNA is a right handed double helix with the chains being complementary and anti-parallel to each other. The two chains are bound to each other with hydrogen bonds between the paired purine/ pyrimidine bases.
The terms 5 prime (5’) and 3 prime (3’) refer to the distinct ends of the chain distinguished by the 5’ phosphate group and 3’ hydroxyl group.
There are major and minor grooves formed from the double helix structure of DNA and the difference between the grooves is the width. As the width of the major groove is wider, the bases are more accessible, therefore proteins that bind to DNA (such as transcription factors) are able to bind.
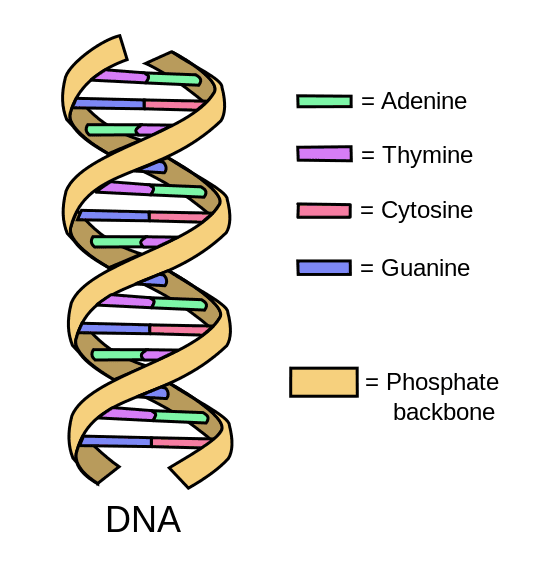
Diagram - The DNA double helix with the base pairings
Creative commons source by Forluvoft [CC BY 3.0 (https://creativecommons.org/licenses/by/3.0)]
The bases on each chain form a hydrogen bond with a specific counterpart on the complementary chain to form a base pair. A purine will always bond with a pyrimidine and vice versa.
In DNA:
- Guanine (purine) forms three hydrogen bonds when it is paired with cytosine (pyrimidine).
- Adenine (purine) forms two hydrogen bonds when it is paired with thymine.
In RNA this is slightly different (pay close attention to how adenosine pairs):
- Guanine (purine) forms three hydrogen bonds when it is paired with cytosine (pyrimidine).
- Adenine (purine) forms two hydrogen bonds when it is paired with uracil.
A way of remembering which base pairs with which can be remembered using animals.
For DNA- GCAT- As G pairs with C and A with T. Imagine a cat with a long tail which curls round to make it look in the shape of a g.
For RNA- GCAU- As G pairs with C and A with U. Imagine a letter G with black and white spots similar to a cow.
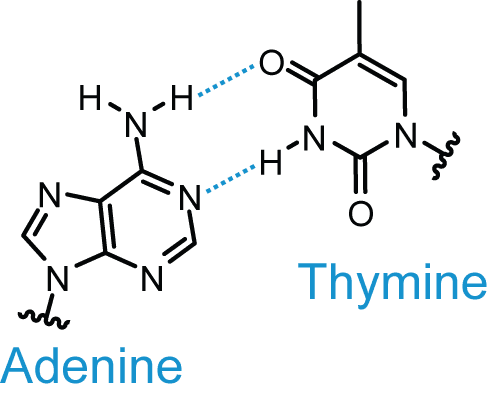
Diagram - The hydrogen bonds between thymine and adenine
Creative commons source by Roadnottaken [CC BY 3.0 (https://creativecommons.org/licenses/by/3.0)]
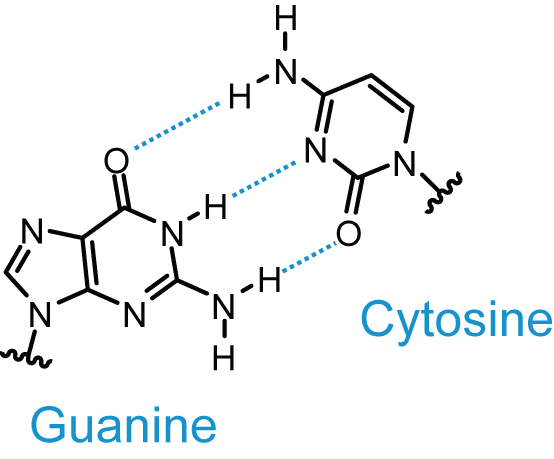
Diagram - The hydrogen bonds between guanine and cytosine
Creative commons source by Roadnottaken [CC BY 3.0 (https://creativecommons.org/licenses/by/3.0)]
Commonly in microbiology, the nitrogenous bases are shortened to the first letter of their name, for example adenine is shorted to A. As well as this, when depicting a DNA sequence, you begin at the 5’ end on the left to the 3’ end on the right.
Replication of DNA occurs in the Synthesis phase (S phase) of the cell cycle (between Growth phase 1 and 2). The reaction is catalysed by the enzyme DNA polymerase. DNA replication is semi conservative in nature and this process is described below.
The existing DNA strands can be described by the direction in which DNA polymerase moves along them. In DNA replication, each original strand acts as a template for the synthesis of a new complementary strand.
DNA polymerase can only work in the 3’ to 5’ direction so therefore produces DNA in the 5’ to 3’ direction (as it produces DNA complimentary to the coding strand i.e. matches the template strand).
There are three stages to DNA replication: initiation, elongation and termination.
- DNA Helicase breaks the hydrogen bonds between the strands to unwind them.
- Primase (type of RNA polymerase) catalyses the synthesis of a short RNA segment (primer) that is complementary to single stranded DNA.
- DNA polymerase cannot begin DNA replication without an initial primer. After elongation, the RNA primer is removed and replaced with DNA by other enzymes before the strand is sealed.
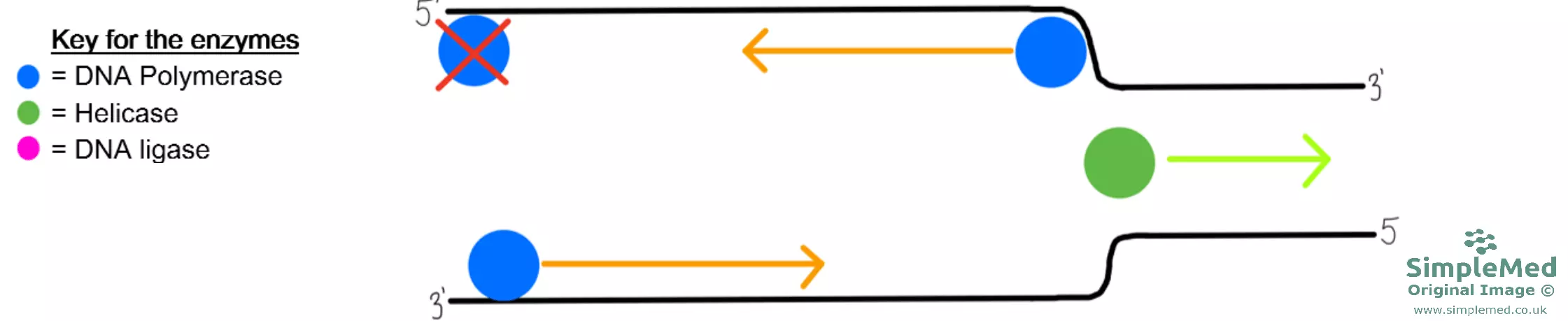
Diagram - The first part of DNA replication showing a replication fork
SimpleMed original by Dr. Peter Parkinson
- On the 3’ to 5’ strand, the same DNA polymerase continues to make a continuous strand of the 5’ to 3’ DNA.
- On the 5’ to 3’ strand, as helicase unwinds the DNA strand, a new DNA polymerase has to begin at the 3’ end leading to multiple DNA polymerase enzymes synthesising new DNA on the 5’ to 3’ strand of the original DNA.
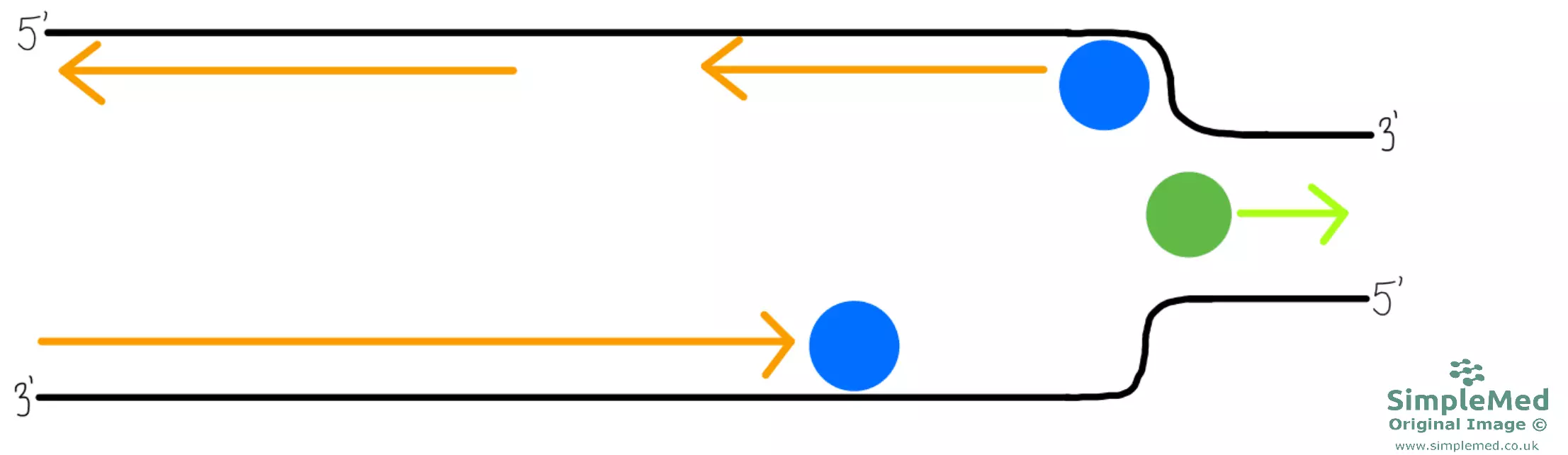
Diagram - DNA replication continued
SimpleMed original by Dr. Peter Parkinson
- With multiple DNA polymerases synthesising new DNA on the 5’ to 3’ strand, there are multiple fragments of new DNA formed. These fragmented segments of new DNA are called Okazaki fragments.
- There are also names for the original strands of DNA depending on whether there are Okazaki fragments or not. 5’ to 3’ strand of original DNA with Okazaki fragments is called Lagging strand whereas 3’ to 5’ strand of original DNA with no Okazaki fragments is called Leading strand.
- These Okazaki fragments on the lagging strands have to be joined up to form a single chain. This is the job of DNA ligase which joins the fragments together.
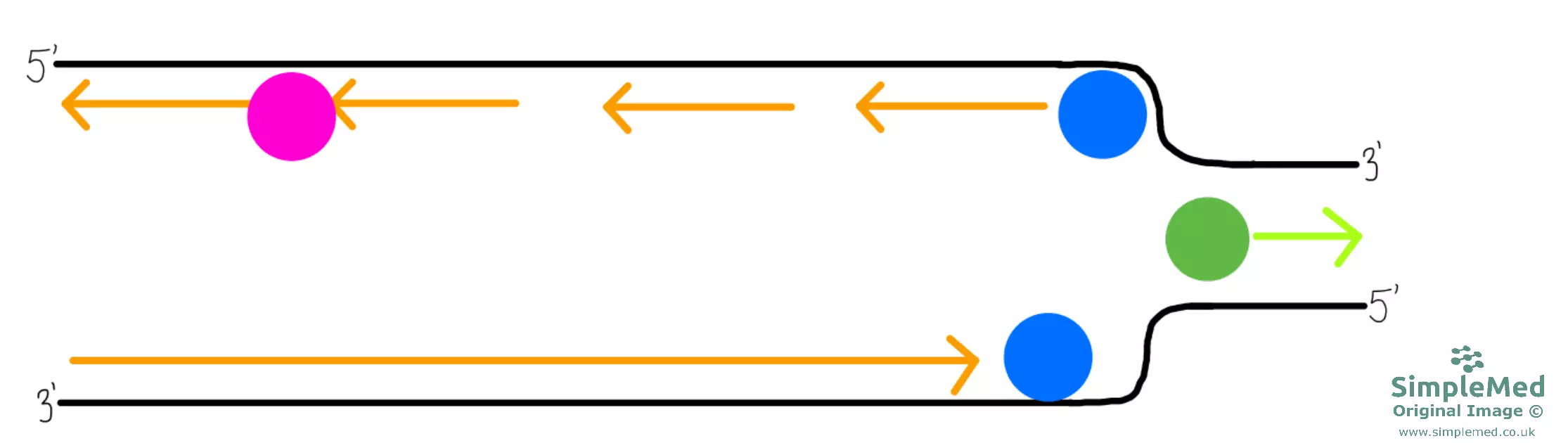
Diagram - DNA replication continued
SimpleMed original by Dr. Peter Parkinson
- As more than one helicase acts on the DNA when replication is required, there are multiple replication forks (sites of replication). These forks with DNA polymerase at the head will of course eventually meet.
- The two remaining Okazaki fragments and the sections at which two forks have met are all joined together by DNA ligase to form two new continuous DNA molecules.
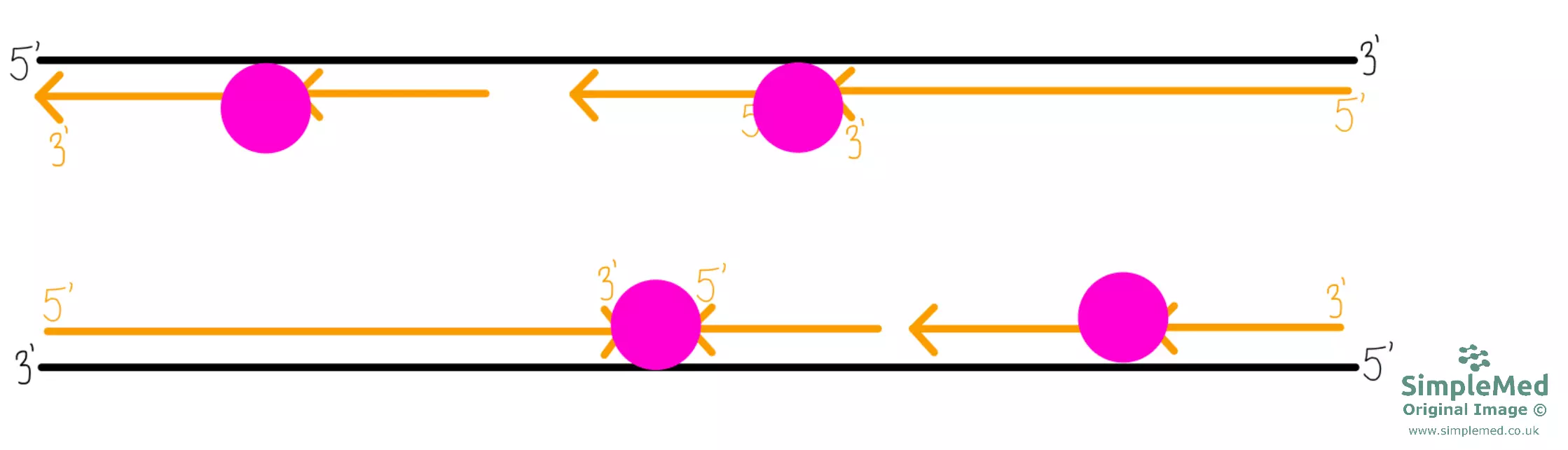
Diagram - DNA replication continued
SimpleMed original by Dr. Peter Parkinson
- After joining of the fragments, there are two new DNA strands contributing to two complete DNA molecules, each with one strand of original DNA and one strand of new DNA. This is why DNA replication is called semi-conservative replication (as half of the original strands are conserved).

Diagram - The end product of DNA replication
SimpleMed original by Dr. Peter Parkinson
DNA replication is a complex process, and the knowledge covered here can be supported by the article in our Genetics series on Mitosis and Meiosis, found here.
Edited by:Dr. Ben Appleby
Reviewed by: Dr. Thomas Burnell
- 10359

