Next Lesson - G-Protein Coupled Receptors
Abstract
- Cell membranes surround the cell and most of its organelles.
- The membrane carries out a variety of different functions.
- Membranes are mainly composed of a phospholipid bilayer and associated membrane proteins.
- The membrane is liquid at room temperature, supporting the Fluid-Mosaic model.
- Membrane fluidity is influenced by temperature, fatty acid saturation and the presence of cholesterol.
- Membrane proteins can be integral or peripheral.
- Proteins of the erythrocyte cell membrane have an important structural role; mutations affecting some of these proteins can lead to hereditary spherocytosis.
- Membrane proteins are initially synthesised into the rough ER membrane.
Core
Cellular membranes are microscopic barriers which form the boundaries of the cell and most of its organelles - they are primarily composed of lipid and protein.
Functions of biological membranes include:
- Acting as a selectively permeable barrier
- Control of the enclosed chemical environment
- Communication and signalling (via receptors and signalling proteins)
- Cell recognition (via recognition of external membrane proteins)
- Cell-cell adhesion (via adhesion proteins)
Cell membranes are composed primarily of a phospholipid bilayer and associated proteins; they also contain a smaller amount of carbohydrate. It is also important to remember that membranes are hydrated structures, with water comprising 20% of the hydrated mass.
Phospholipids are amphipathic molecules, meaning that they have both hydrophilic and hydrophobic parts. This causes them to form a bilayer when exposed to water, with hydrophobic tails pointing inwards and hydrophilic heads facing outwards, in contact with water.
The hydrophobic portion consists of 2 hydrocarbon tails (derived from fatty acids), bound to a molecule of glycerol. The fatty acid tail can be saturated or unsaturated - saturated fatty acids have all single C-C bonds, whereas unsaturated fatty acids have at least one C=C double bond (cis configuration).
Also bound to the glycerol is a phosphate head, which makes up the hydrophilic portion of the phospholipid. The phosphate head is often modified with another organic molecule: these include choline (most common), serine, inositol and ethanolamine.
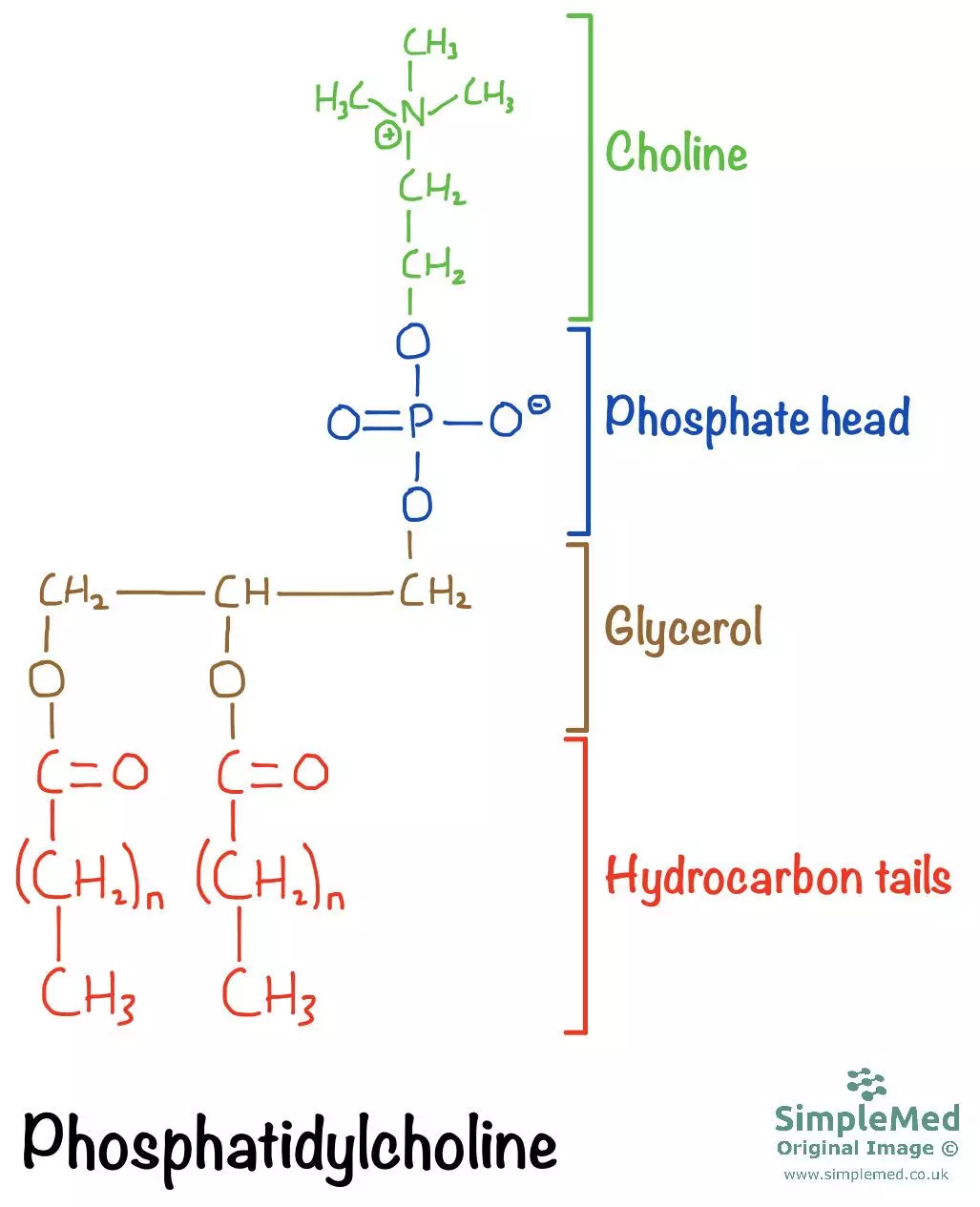
Diagram - Chemical structure of the phospholipid Phosphatidylcholine
SimpleMed Original by Dr. Josh Bray
Glycolipids are modified lipids which contain a carbohydrate group instead of a phosphate group. These can be classified as either cerebrosides, which possess a simple sugar monomer, or gangliosides, which possess a longer oligosaccharide (multiple sugar units).
The membrane is generally in a liquid state at room temperature, depending on lipid composition. This supports the idea of a Fluid-Mosaic model, which describes a fluid lipid bilayer with interspersed proteins which are free to move throughout the bilayer.
Phospholipids are capable of a wide range of movement:
- Flexion - Changing the angle between hydrocarbon tails
- Rotation
- Lateral diffusion - Movement throughout its own monolayer
- Flip-flop (rare) - Highly energetic process where the phospholipid swaps to the other side of the bilayer
Membrane proteins are a bit more restricted in terms of mobility:
- Conformational change - Changes in shape (important for function)
- Rotation
- Lateral diffusion
Proteins are not capable of flip-flop as they are too large and so would require too much energy for spontaneous flip-flop.
Many factors contribute to the fluidity of the membrane:
- Temperature (normally constant) - As temperature increases, membrane fluidity increases due to increased kinetic energy.
- Unsaturated fatty acids - C=C double bonds introduce a ‘kink’ in the hydrocarbon tail, increasing the spacing between neighbouring phospholipids and therefore increasing membrane fluidity.
- Cholesterol - Acts as a stabilising agent due to its paradoxical effects on membrane fluidity; its rigid (planar) sterol ring reduces fluidity whereas its flexible hydrocarbon tail increases fluidity.
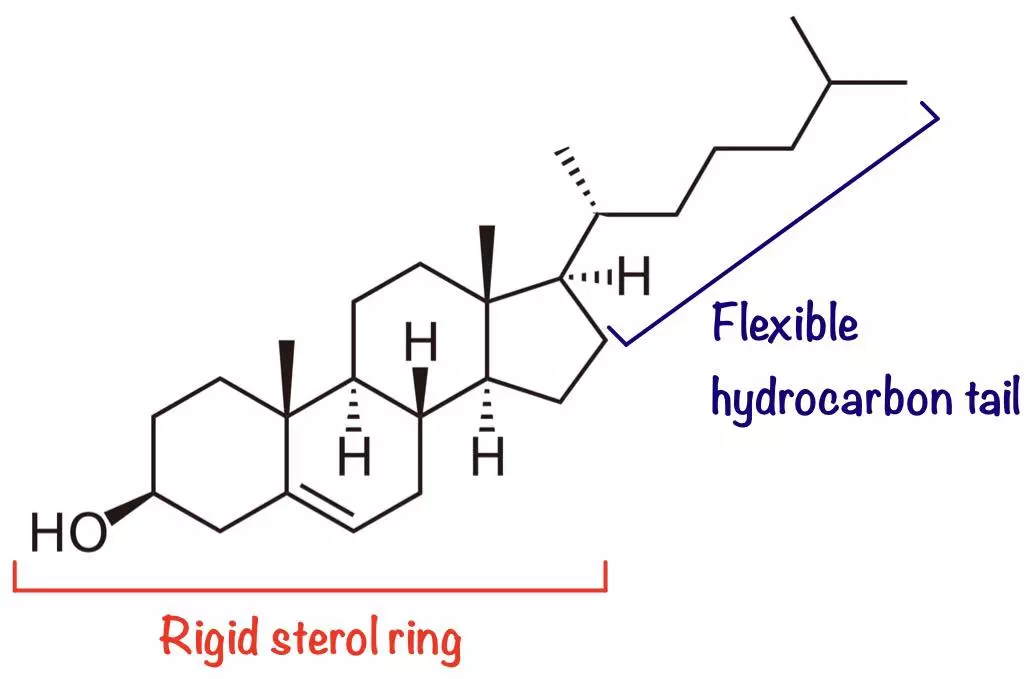
Diagram - Chemical structure of cholesterol
Public Domain Source by BorisTM [Public domain], edited by Dr. Josh Bray
Control of membrane fluidity is important as it has implications for structure and membrane permeability - the more fluid the membrane, the higher its permeability.
Despite in theory being able to move freely through the lipid bilayer, the movement of membrane proteins can be restricted by several mechanisms:
- Tethering - To the cytoskeleton or extracellular matrix (particularly the basement membrane).
- Aggregation - Proteins with complementary chemical and physical properties will tend to stay close to one another.
- Cell-cell adhesion - Via cadherins, desmosomes, tight junctions and gap junctions.
Tight junctions play a particularly important role in cell polarisation, as they separate protein populations on the apical and basolateral membranes.
Proteins associated with the cell membrane can be integral (intrinsic) or peripheral (extrinsic):
Integral proteins are embedded within the phospholipid bilayer; they may or may not span the entire width of the membrane (where they are referred to as transmembrane). These proteins must have an extensive hydrophobic domain to interact with the hydrocarbon tails of the lipid bilayer. Examples include channels, transporters, receptors (namely G-protein coupled and tyrosine kinase receptors), adhesion proteins and some enzymes (e.g. the Na+/K+ ATPase).
Peripheral proteins are bound to the surface of the membrane via electrostatic and hydrogen bond interactions with the polar head groups of phospholipids. Unlike integral proteins, peripheral proteins can become dissociated from the membrane by disturbances in pH as this will change the ionic configuration of amino acid residues. Examples include G-proteins, cytoskeletal proteins (e.g. spectrin), electron carriers (e.g. cytochrome C) and some enzymes (e.g. phospholipase C).
Membrane proteins can also be modified with a carbohydrate group to form glycoproteins.
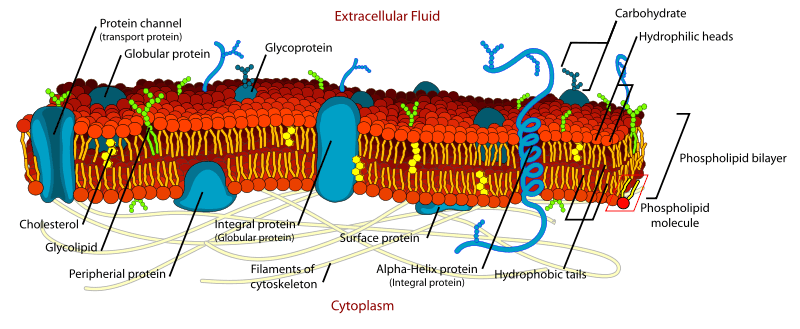
Diagram - Structure of the cell membrane, showing the different types of membrane proteins
Creative commons source by Mariana Ruiz, edited by Alokprasad84 [CC BY-SA 4.0 (https://creativecommons.org/licenses/by-sa/4.0)]
Proteins of the erythrocyte membrane are essential for maintaining the biconcave shape of the cell. This is achieved by vertical interaction between the membrane, its associated membrane proteins and the cytoskeleton.
The proteins of the erythrocyte membrane include:
- Band 3 - A Cl-/HCO3- exchanger (integral protein) which links to protein 4.2 and ankyrin.
- Ankyrin - Links integral membrane proteins to the underlying spectrin-actin cytoskeleton.
- Protein 4.2 - ATP-binding protein which may regulate the association of band 3 with ankyrin.
- Spectrin - Crosslinks with actin, forming a molecular scaffold that links the plasma membrane to the actin cytoskeleton.
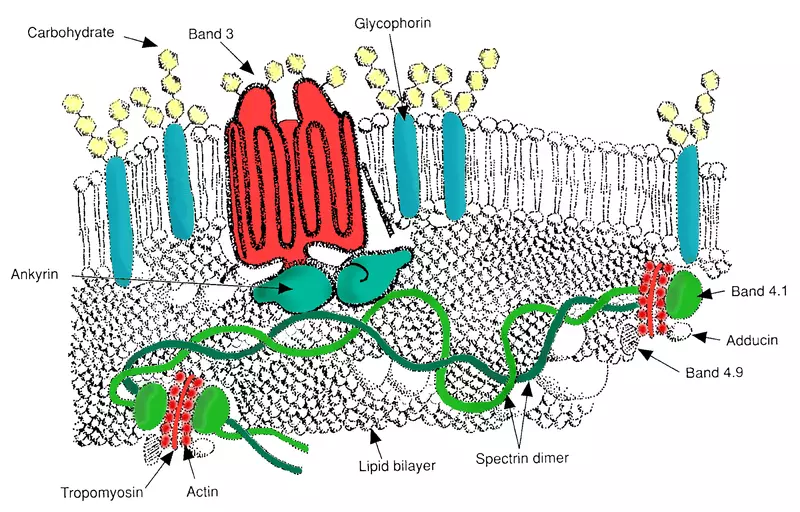
Diagram - Structure of the erythrocyte membrane, with major proteins included
Public Domain Source Ernst Hempelmann [Public Domain]
Hereditary Spherocytosis
Deficiency of the functional form of any one of these proteins (most commonly ankyrin) can result in a condition called hereditary spherocytosis. This is a condition where red blood cells take on a spherical shape instead of the classical biconcave shape, due to loss of vertical interaction between the cytoskeleton and plasma membrane. This causes erythrocytes to become less deformable, leading to haemolysis.
Proteins destined for the cell membrane are synthesised into the membrane of the rough endoplasmic reticulum, where they can bud off as a vesicle and fuse with the cell membrane.
The processes involved in membrane protein biosynthesis are as follows:
- A ribosome in the cytosol begins the process of translation from a template molecule of mRNA.
- A signal recognition particle (SRP) recognises the intrinsic signal sequence in the growing polypeptide chain and binds to it, bringing protein synthesis to a halt temporarily.
- The SRP-ribosome complex moves to the rough ER at the site of an SRP receptor (docking protein).
- SRP dissociates from the ribosome and binds to the SRP receptor for recycling, while the ribosome forms a translocation complex (translocon) with the help of ribophorins in the ER membrane.
- Protein synthesis continues and the polypeptide grows into the ER lumen.
- The signal sequence on the polypeptide is cleaved by a signal peptidase within the ER lumen.
- A specific hydrophobic portion of the polypeptide (stop-transfer sequence) becomes embedded within the ER membrane and protein synthesis is completed by the ribosome.
- The ribosome detaches, leaving the protein embedded within the membrane.
NB: The process for peripheral membrane proteins will be slightly different as they do not become embedded in the membrane.
Reviewed by: Dr. Marcus Judge
- 11255

