Next Lesson - Gene Expression
Core
Our genotype, put simply, is the sum total of all our genes, encoded by DNA. Our phenotype is the physical manifestation of our genotype via proteins. For example, the human blood types are encoded by the ABO gene found on chromosome 9 (genotype). When translated the gene produces glycoproteins on the surface of red blood cells (phenotype).
It is important to note that the environment has an important effect on our phenotype. For example, Familial Adenomatous Polyposis genotypically predisposes someone to developing bowel cancer, but additional genetic damage influenced by environmental factors is required for cancer to develop. Similarly, the BRCA1 gene greatly increases a woman's lifetime risk of developing breast cancer to around 65-85%. However, as this risk is not 100% (that is, BRCA1 does not always lead to cancer), non-genetic factors must also be involved.
Each human has 25,000 genes, with two copies of each gene. The two copies of a gene may be the same or through random mutation may be ever so slightly different. Variants of the same gene are known as an allele. As we inherit one half of our genetic information from our mother, and one half from our father we inherit two unique sets of genes, and for a given gene we may inherit different alleles.
For a particular gene, an individual can be described as homozygous (both the allele from the mother and the father are the same) or heterozygous (both alleles are different). For the third category, hemizygous (only one allele of a gene is present on the x chromosome), it is important to understand that only males can be hemizygous, as they only have one x chromosome. Females cannot have a single copy of an allele for an X chromosome as they have two X chromosomes.
In a heterozygote, the dominant allele determines the phenotype, and the non-dominant allele does not determine the phenotype and is called recessive.
In some cases, for example the ABO gene, it is possible for two different dominant alleles in a heterozygote to exert an effect on the phenotype; this is known as co-dominance.
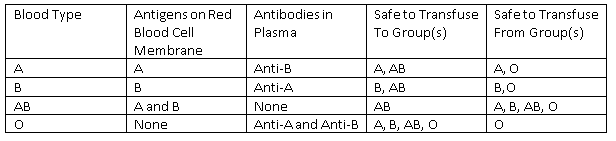
Figure: Blood Group Transfusion Table
SimpleMed original by Dr. Joshua Sturgeon
Please note, in reality a protein known as Rhesus Factor also plays a role in determining a person’s blood type (a person can be Rhesus Factor positive (+) or negative (-)), but this is beyond the scope of this article (see our haematology section). Just appreciate that blood group O- is known as the ‘universal donor’ as it is safe to transfuse into any patient, so is used in an acute situation where the patient’s blood group is unknown.
There are three alleles of the ABO gene: IA, IB, and IO. IA is dominant over IO. IB is also dominant over IO. However, neither allele IA or IB is dominant over the other, they are co-dominant. This co-dominance allows us to have the blood type AB. IO is the only allele of the ABO gene which is recessive.

Figure: Possible Genotypes for Blood Groups
SimpleMed original by Dr. Joshua Sturgeon
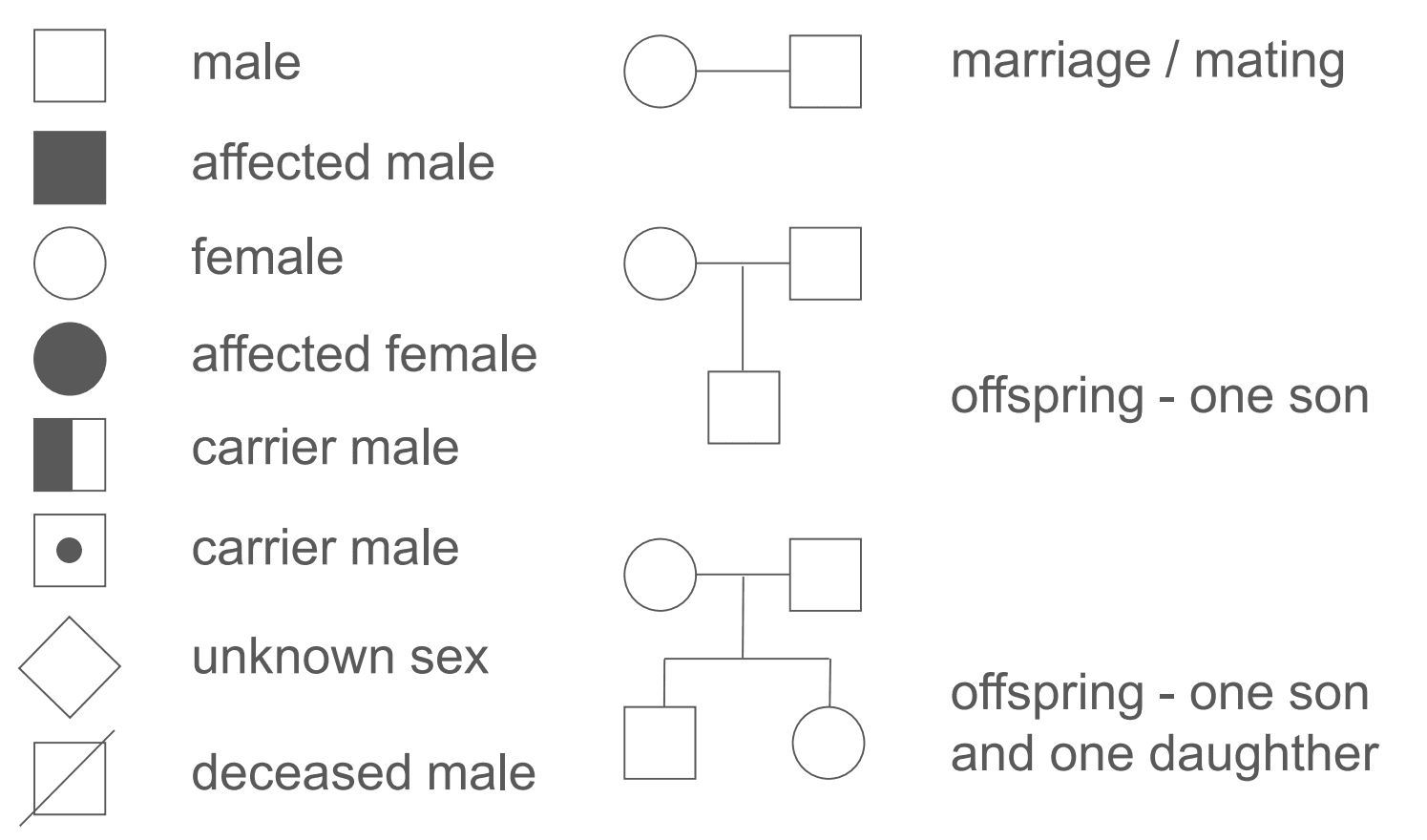 Figure: Pedigree Key
Figure: Pedigree Key
SimpleMed original by Dr. Joshua Sturgeon
Pedigrees are drawn using the symbols shown above. Each generation is placed on a separate line, each of which is numbered with a roman numeral. When drawing a generation of offspring, it is convention to have the oldest offspring on the left hand side. Within a generation, each individual is numbered from left to right. An example on how to interpret a pedigree chart is shown below.
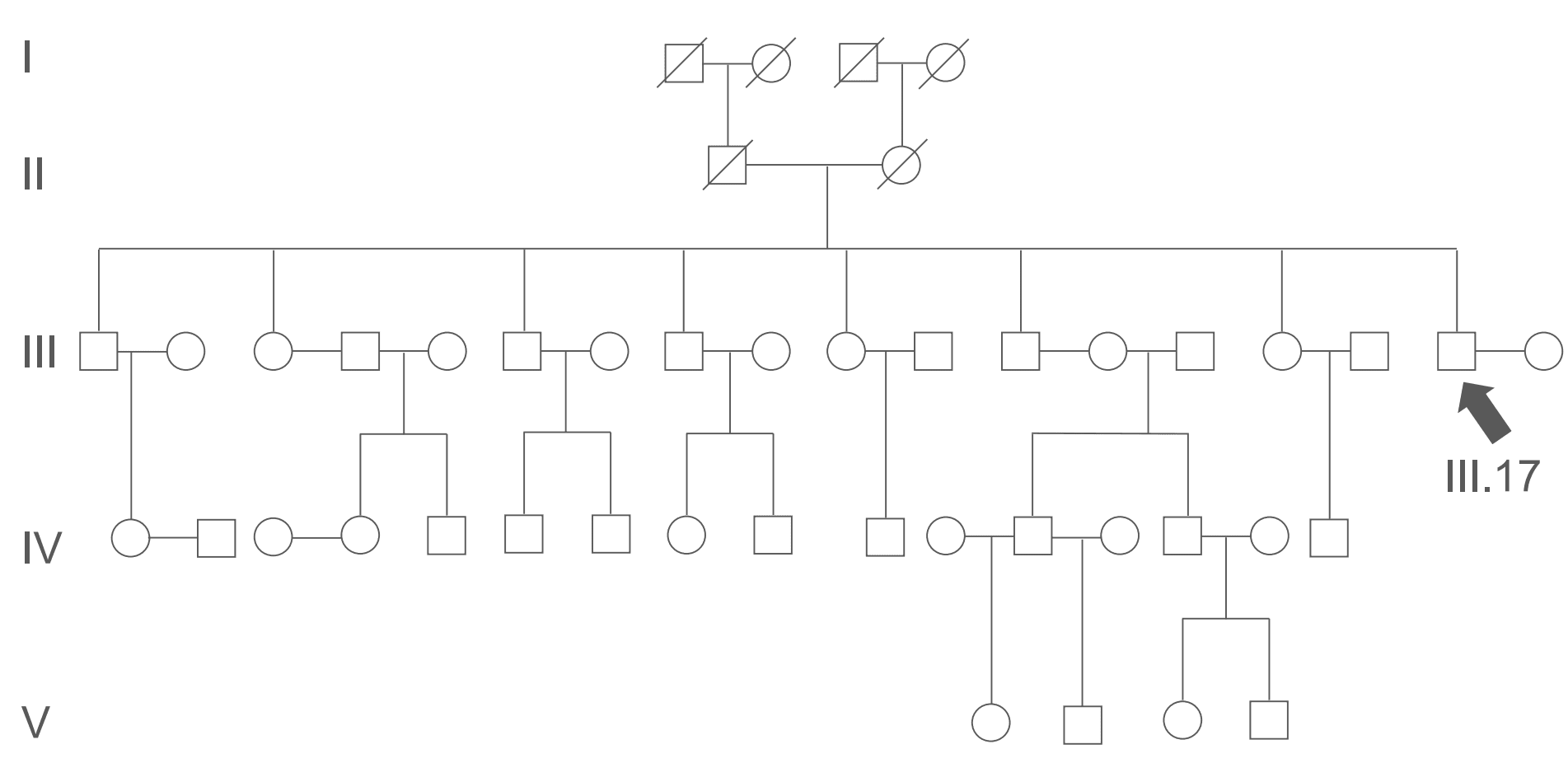
Figure: Example Pedigree Chart. All members of generations I and II are deceased. The relations of III.17 are: wife III.18, parents II.1 and II.2, a sister III.3, and a niece IV.1. III.17 is genetically unrelated to III.4 and III.5, and to III.13 and III.14, and is therefore genetically unrelated to their respective offspring.
SimpleMed original by Dr. Joshua Sturgeon
Being able to interpret or even draw a pedigree chart from a given family history is a useful skill to have. An understanding of determining an inheritance pattern of an unknown disease from a pedigree chart is crucial.
- Autosomal (carried on chromosomes 1-22 i.e. non sex chromosomes)
- Autosomal Recessive
- Autosomal Dominant
- Sex-linked
- X-linked Recessive
- X-linked Dominant
- Y-linked
- Mitochondrial (inherited through mitochondrial DNA)
- Polygenic/Complex or Multifactorial
We will go into explanations for the above inheritance patterns, along with Punnett squares and pedigree charts below.
For an individual to be affected with an autosomal recessive disease, they must be a homozygous recessive (carrying two recessive alleles for the disease gene). Heterozygotes are unaffected as they have a dominant allele, but they are known as carriers. If two heterozygous carriers reproduce, each parent has a 50% chance of passing on the recessive allele, giving a 25% chance that the child will be affected. Two affected individuals for a particular disease (homozygotes) will only ever have affected offspring. Males and females are equally affected. An example of an autosomal recessive disease is Cystic Fibrosis.
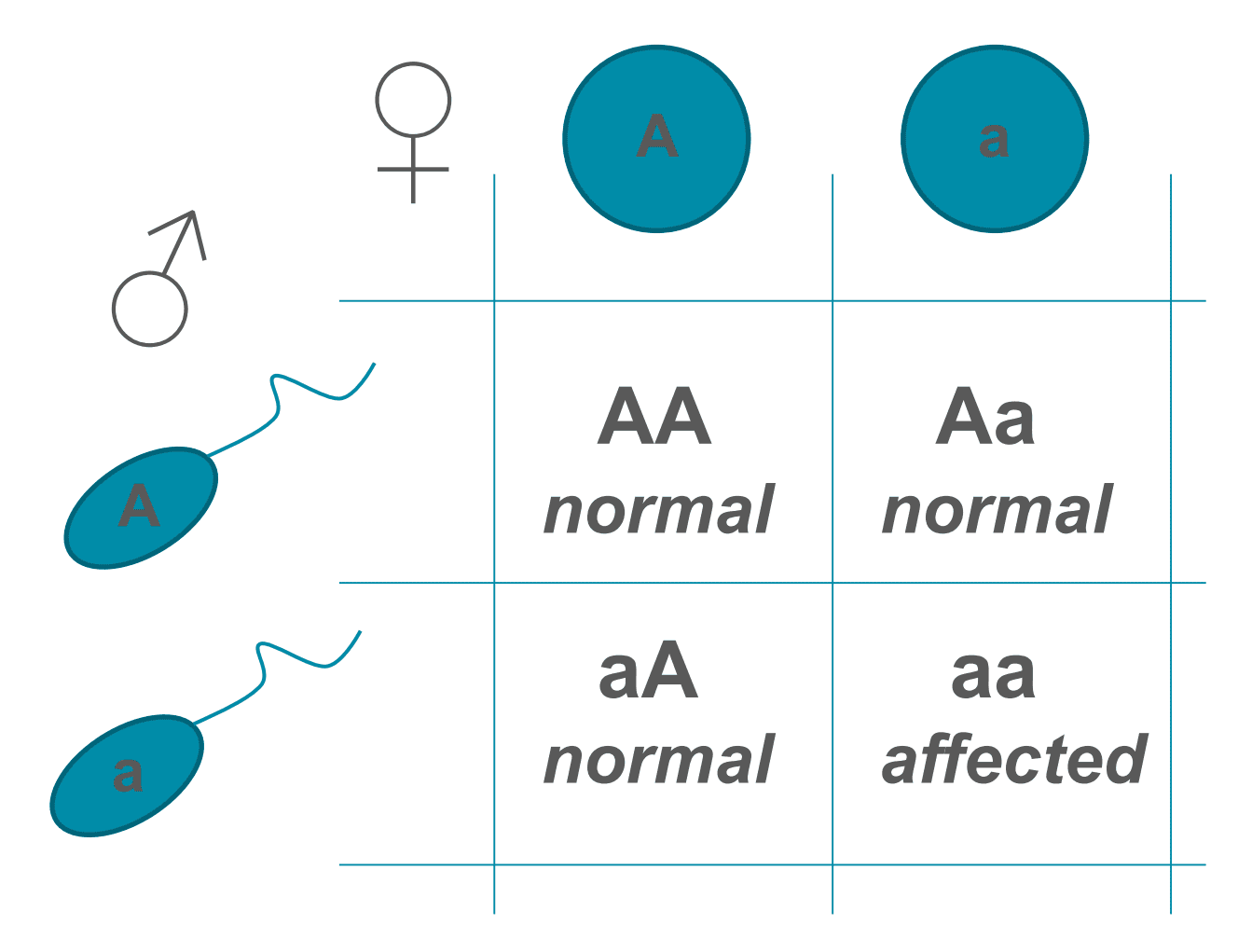
Figure: A punnet square showing autosomal recessive inheritance with two heterozygotes. Note that there is a 25% chance of the offspring being affected by the disease
SimpleMed original by Dr. Joshua Sturgeon
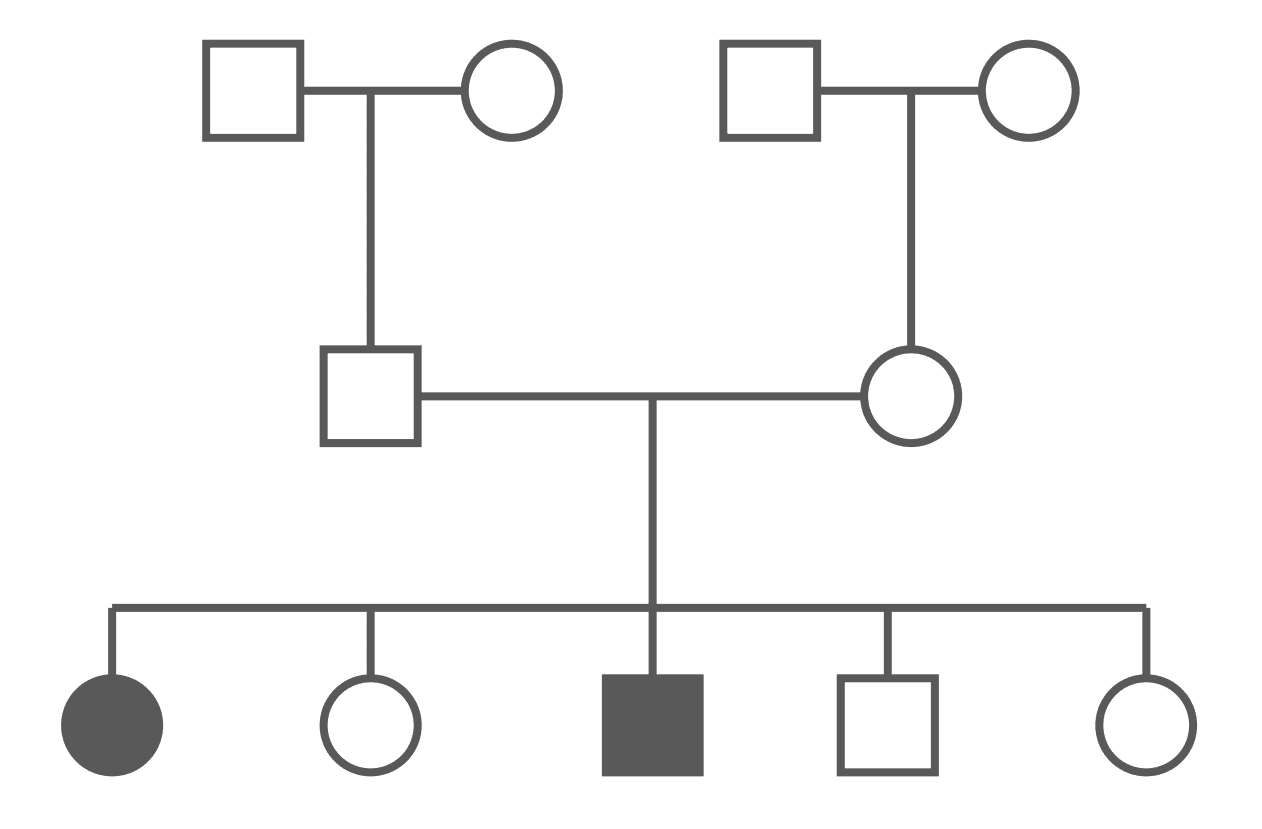
Figure: Autosomal Recessive Pedigree Chart (Carriers Not Shown)
SimpleMed original by Dr. Joshua Sturgeon
An autosomal recessive disease can appear to a patient’s family to ‘come out of nowhere’. The disease can also skip generations. It is important to note that whilst there are icons for carriers in the key above, a pedigree may not always show carriers (as many people will not know they carry the gene and as such will not be included in a history), which makes it a little harder to determine the pattern.
For a person to suffer from an autosomal dominant disease, they only need one allele (heterozygous). The disease is rarely found in a homozygous state, potentially due to the increased morbidity associated with being a homozygote for an autosomal dominant disease. Every affected individual has a 50% chance of having an affected offspring. Males and females are affected equally. An example of an autosomal dominant disease is Huntington’s Disease.
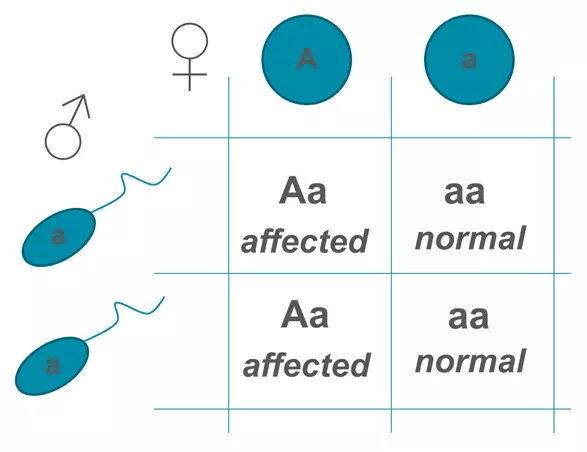
Figure: Autosomal Dominant Punnet Square
SimpleMed original by Dr. Joshua Sturgeon
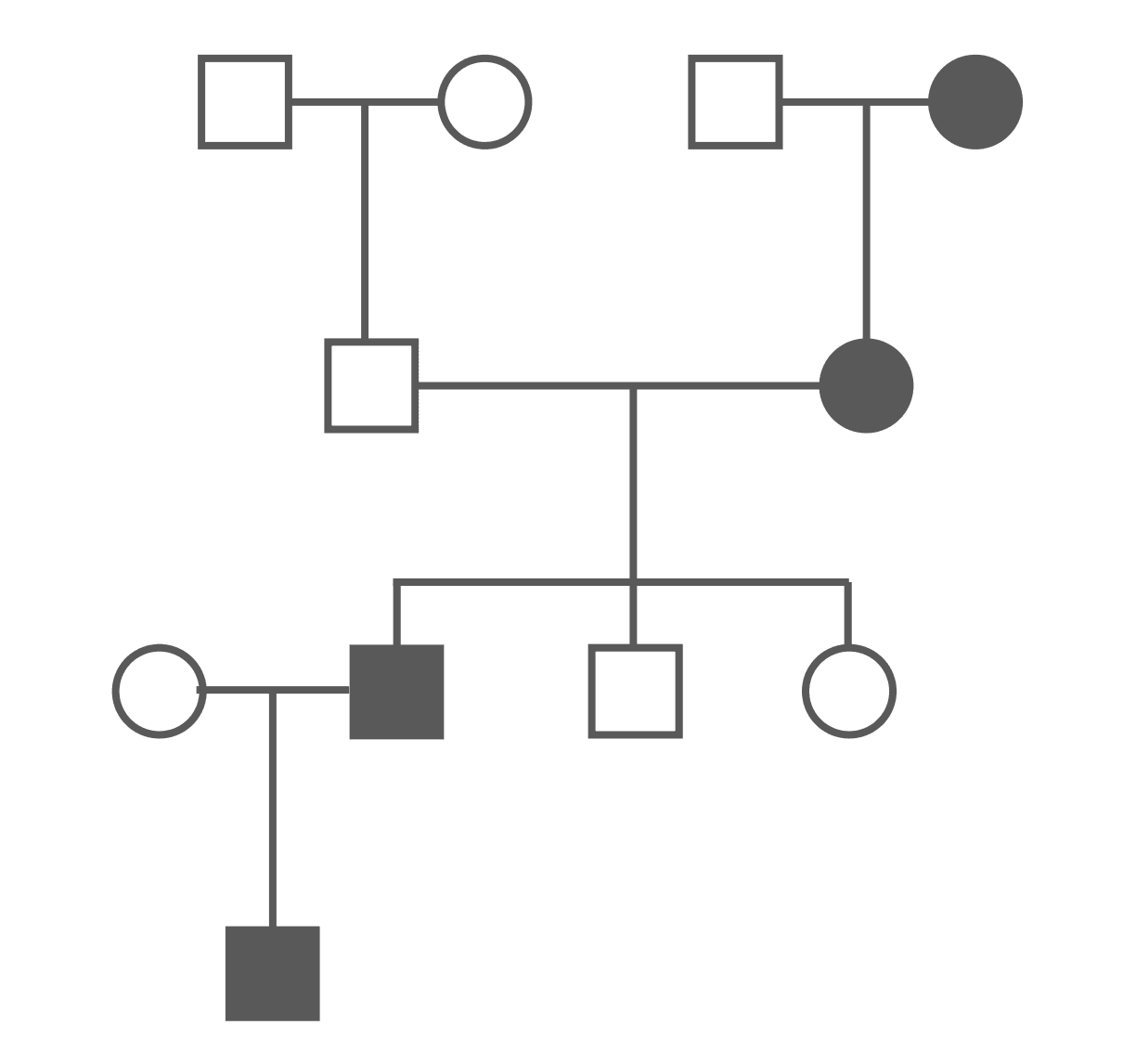
Figure: Autosomal Dominant Pedigree Chart
SimpleMed original by Dr. Joshua Sturgeon
One of the major differences between autosomal recessive and autosomal dominant in a pedigree chart is that autosomal dominant conditions usually affect successive generations. In some cases, reduced penetrance can make a condition appear to skip a generation.
An important point to note is that for an autosomal dominant condition to be inherited (like any disease) the offspring must survive to reproductive age. For example, if an autosomal dominant condition is lethal by the age of 5 it can’t be inherited. Huntington’s is notable as although it is lethal, symptoms do not usually present until the patient is at least 30 years of age. This means the patient is old enough to have had children and will have passed on the disease.
X-linked diseases refer to diseases for which genes are carried on the X chromosome. Therefore, females (XX) must be homozygous for the recessive alleles however, a male must only be hemizygous. As males have one X and one Y chromosome, males are much more likely to be affected by these diseases. A heterozygous female carrier has a 50% chance of having affected sons, whereas affected males cannot give the trait to their sons, only their daughters, as a son would inherit the Y chromosome from their father, and the X chromosome from their mother. An example of an X-linked recessive disease is Haemophilia A.
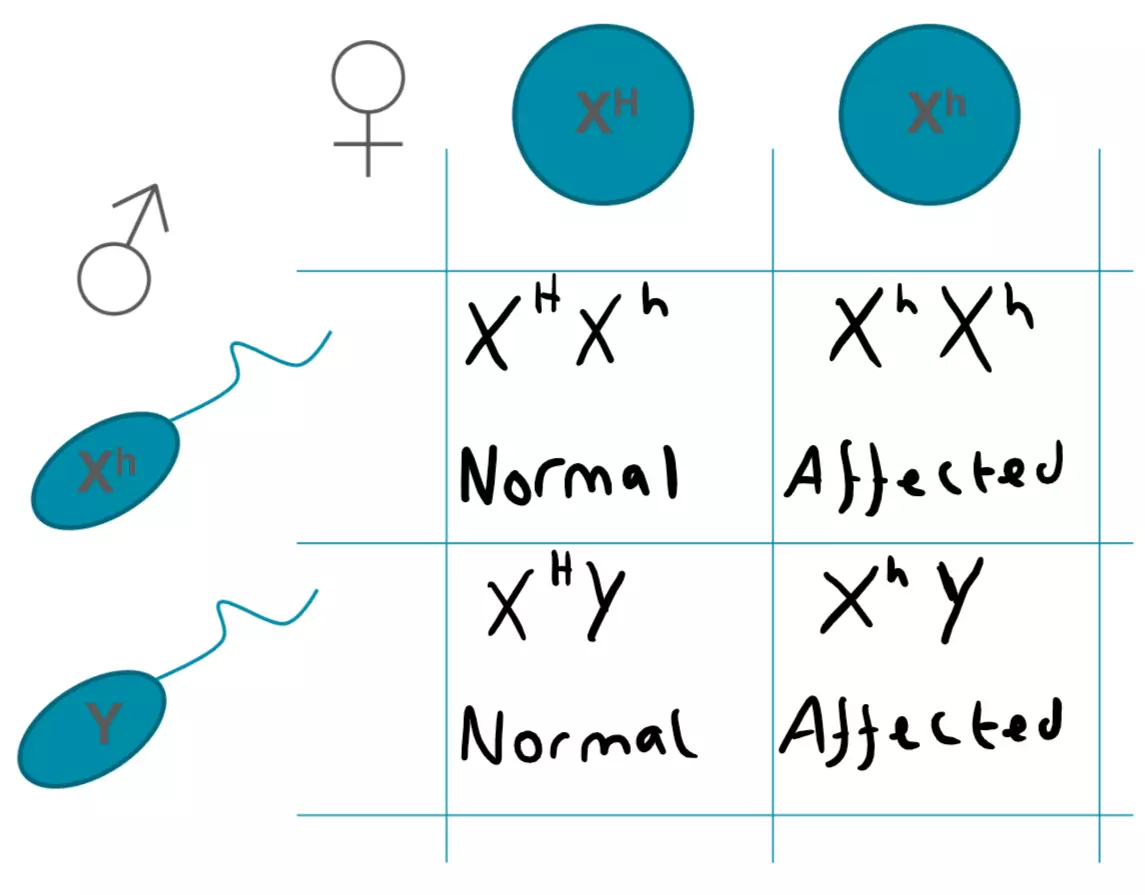
Figure: X-linked Recessive Punnet Square
SimpleMed original by Dr. Joshua Sturgeon
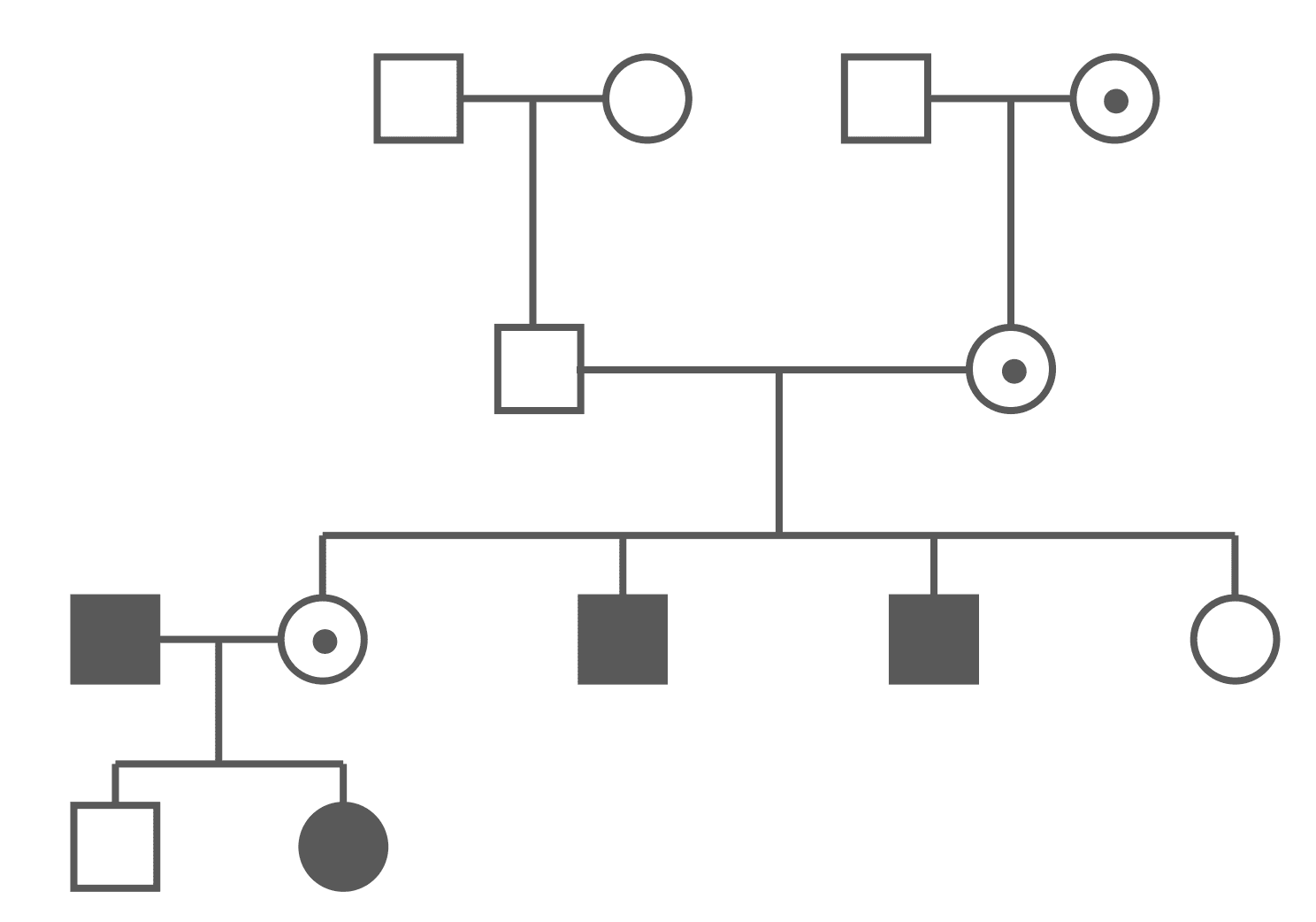
Figure: X-linked Recessive Pedigree Chart (Carriers Shown)
SimpleMed original by Dr. Joshua Sturgeon
The above pedigree chart shows that males and females are unequally affected. Every affected male must have at least a carrier mother. Every affected female must have an affected father and a carrier (or affected) mother.
X-linked dominant diseases affect hemizygous males and heterozygous females. A heterozygous female has a 50% chance of having affected offspring, whereas an affected male cannot give the trait to his sons but will give it to all his daughters. X-linked dominant diseases are uncommon compared with autosomal inheritance patterns. An example is incontinentia pigmenti, a progressive dermatological disorder affecting the skin, hair, teeth, and central nervous system.
Exam tip: X linked conditions are extremely rare. Therefore, although it is important to be familiar with their inheritance pattern, when faced with analysing a pedigree try to see if more common inheritance patterns (see above) fit before reaching for rarer patterns. 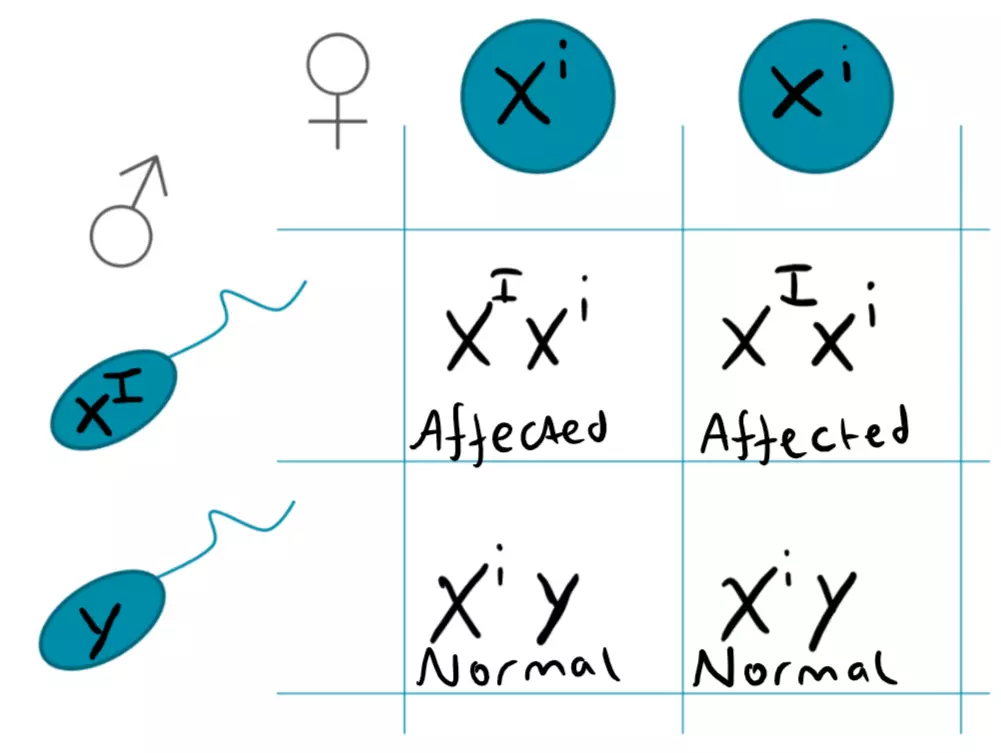
Figure: X-linked Dominant Punnet Square
SimpleMed original by Dr. Joshua Sturgeon
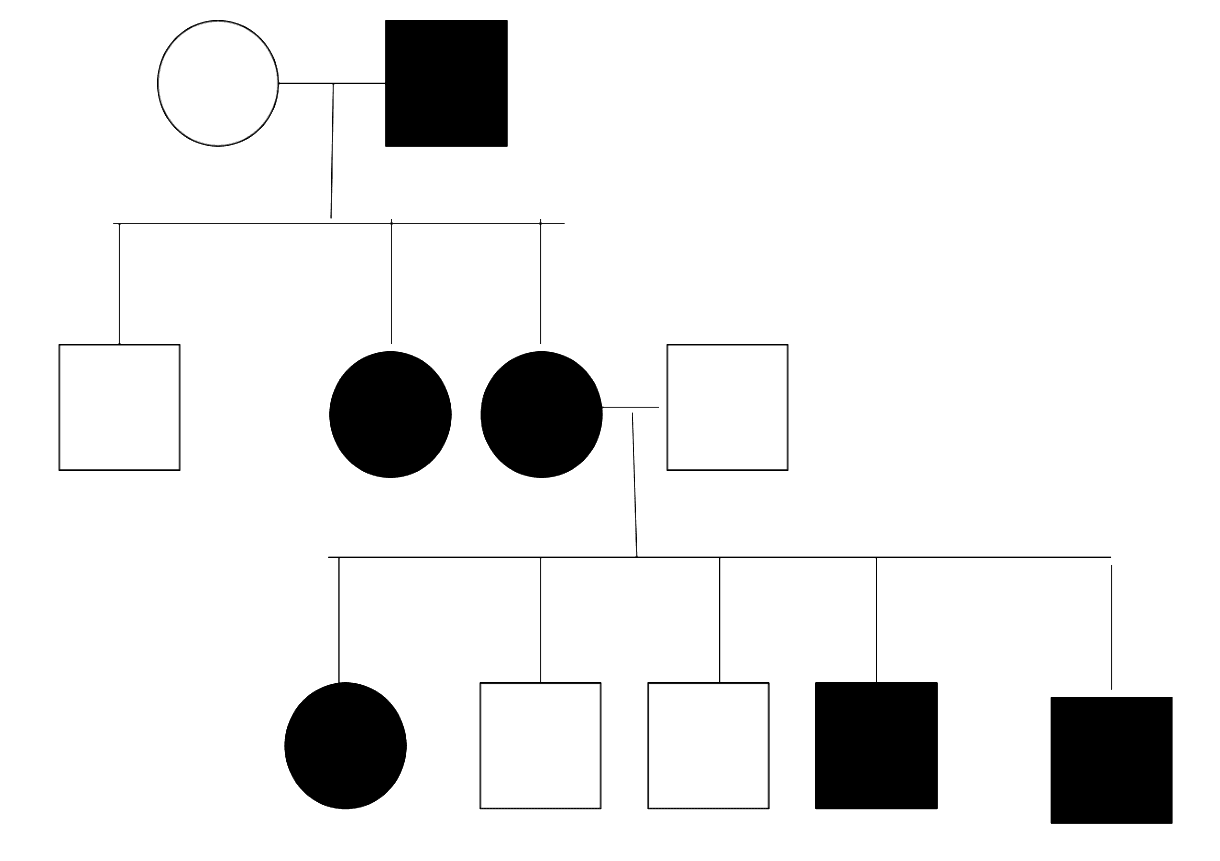
Figure: X-linked Dominant Pedigree Chart
Creative commons source by Hydra2114 [CC BY-SA 4.0 (https://creativecommons.org/licenses/by-sa/4.0)]
As males are the only gender to have the Y chromosome, y-linked diseases only affect males. The y chromosome contains genes which drive normal male development, such as the SRY gene. As there is only one Y chromosome in males, Y-linked traits are usually described without using dominant or recessive terminology, as any functional allele present is expressed. It is worth noting that y-linked diseases are even rarer than x-linked dominant diseases, so it can be hard to find more information about them in medical literature. An example of a y-linked disease is Azoospermia which is the complete inability to produce sperm, leading to infertility.
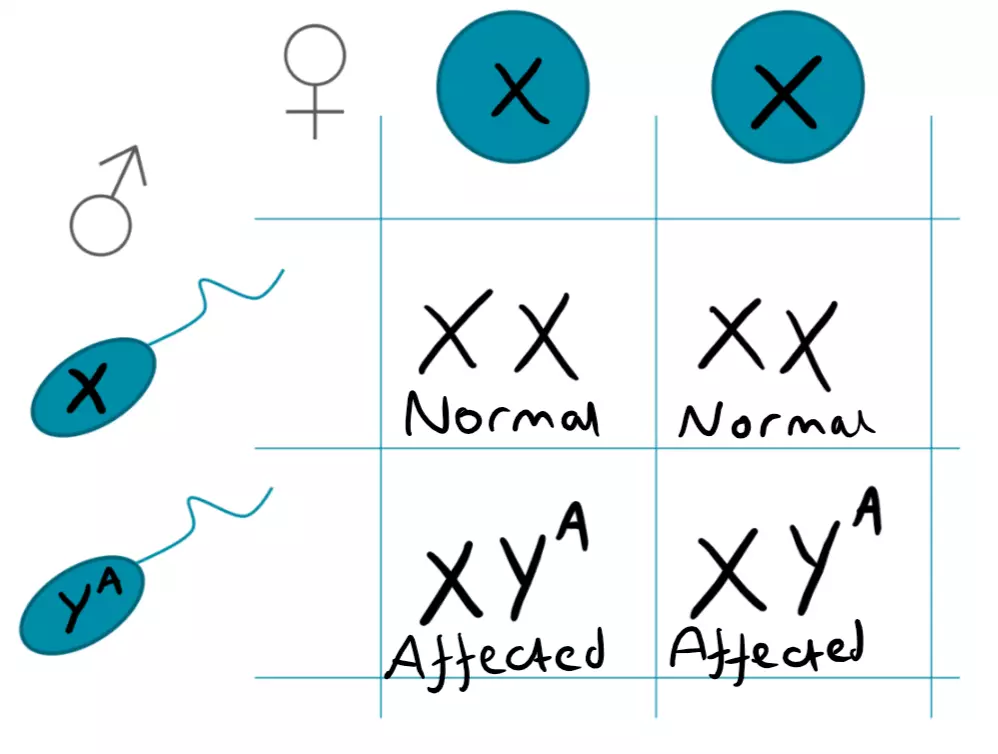
Figure: Y-linked Punnet Square
SimpleMed original by Dr. Joshua Sturgeon
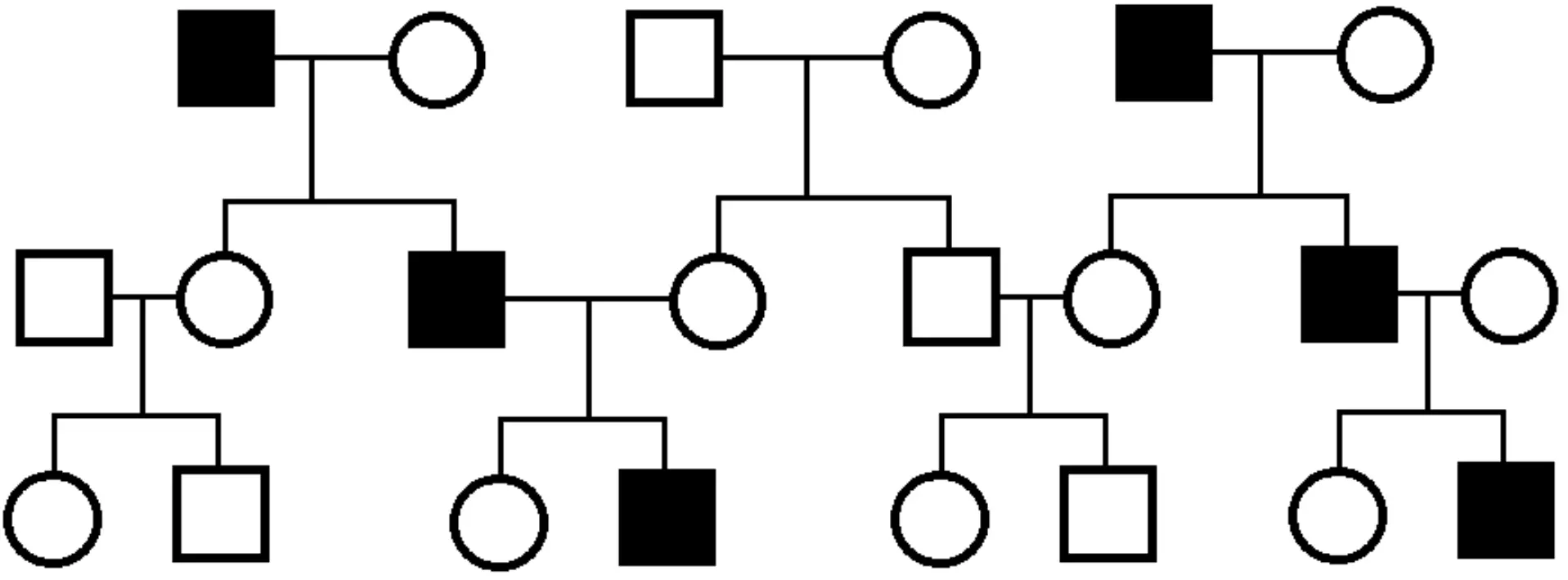
Figure: Y-linked Pedigree Chart
Public Domain Source
Mitochondria contain DNA (mtDNA) which is used to encode proteins for the oxidative phosphorylation step of respiration. mtDNA is only inherited from the mother, as the mitochondria of the egg produce all the mitochondria for the cells of the body. A mutation in mtDNA will cause malformation of the proteins required to generate the large amount of ATP needed by cells, so will lead to mitochondrial diseases.
A new and exciting technique known as ‘three parent babies’ has been used to combat inherited mitochondrial diseases. https://www.bbc.co.uk/news/health-37485263
So far, we have only discussed traits which are caused by one gene. However, in reality more than one gene is usually involved in producing a phenotype, such as height or skin colour, as is shown below.
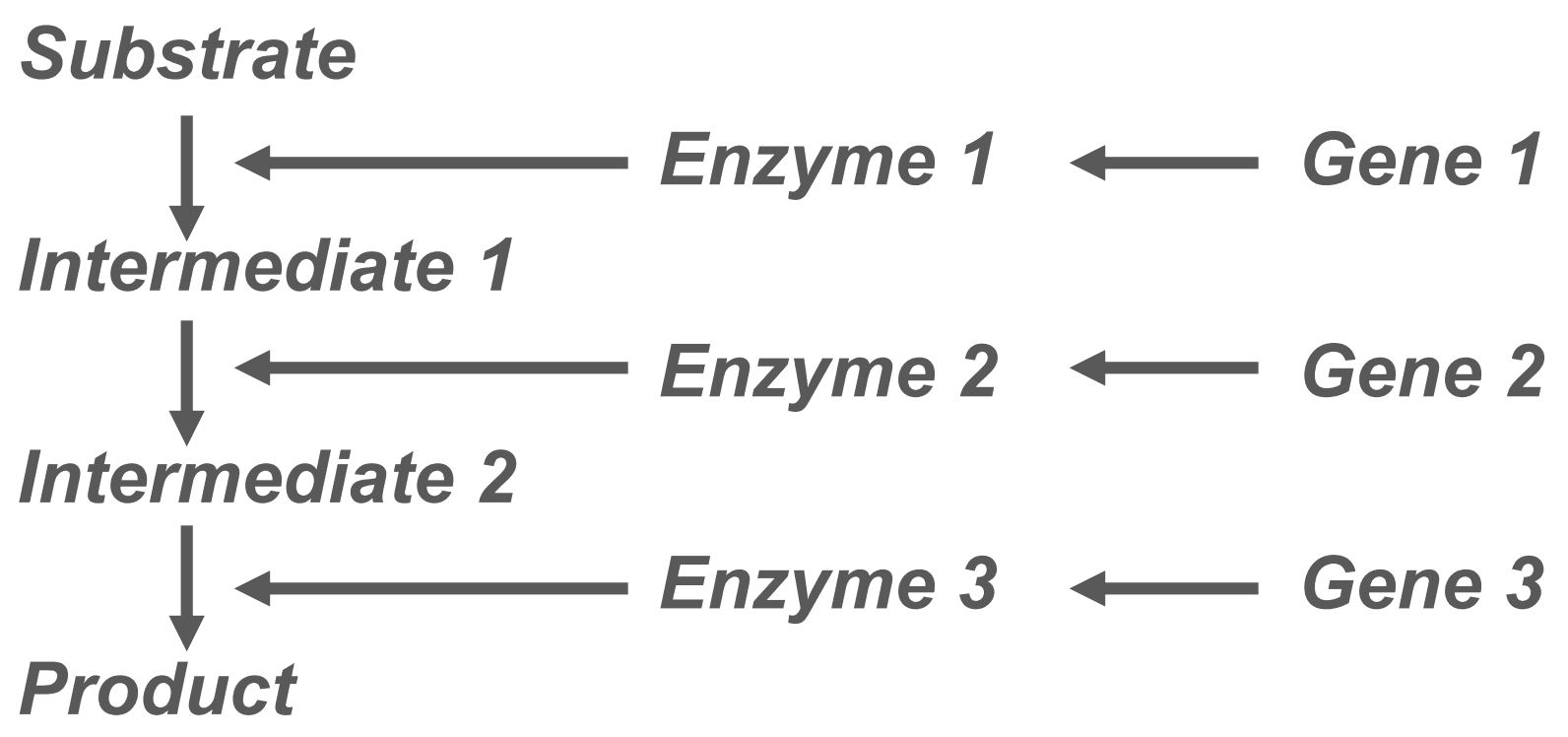
Figure: To demonstrate how multiple genes can have an influence on one phenotype
SimpleMed original by Dr. Joshua Sturgeon
Gene linkage is the process by which genes which are close together on the same chromosome are inherited together. Genes are ‘linked’ when they are on the same chromosome, especially if they are close together. This is as during crossing over, parts of the chromosome can be swapped with its homologous partner. Genes which are closely situated to each other on the chromosome are less likely to be separated by recombination and are therefore ‘linked’ as they are more likely to be inherited together. Genes on separate chromosomes or which are far apart on the same chromosome are not linked as they are inherited independently or are separated during crossing over.
If you are struggling to understand gene linkage, don’t worry, just remember that gene linkage means it is difficult to predict the genotypes of offspring when the genes involved are located close together on the same chromosome.
Exam Tip:
If a question about inheritance is difficult, and you are struggling to filter down the information, try to highlight the genotypes or phenotypes given in the stem, and use them to draw a Punnett square. This will allow you to easily calculate the probabilities of different offspring genotypes. This approach can even be used if you are asked to interpret a pedigree chart and determine the pattern of inheritance.
Another good rule to remember is that if the question refers to inheritance of a dominant condition but does not explicitly state the genotypes of the parents, assume that the parents' genotypes are heterozygous.
Reviewed by: Dr. Marcus Judge
- 14072

