Next Lesson - Introduction to Gene Mutation
Abstract
- Gene Expression is the process of converting DNA -> mRNA -> Protein, and involves transcription and translation.
- Transcription converts the information encoded on genes into mRNA, which is the basis for building a protein.
- Translation occurs at ribosomes and uses mRNA as a template. tRNA brings amino acids to the ribosome in order to build a polypeptide (protein).
Core
Gene expression is the process which involves transcription of DNA to messenger RNA (mRNA), and then the translation of mRNA to a protein.
A gene is a stretch of DNA, which acts as the ‘recipe’ for a protein. Each gene has a chromosomal locus - e.g. the CFTR gene has a locus of 7q31.2 - and while this may look complicated, it simply tells you the physical location of the gene (CFTR is on the q arm of chromosome 7, at location 31.2). At first, it may seem useless to give each gene a locus, but there are two copies of around 20,000 genes per diploid cell, so we need a system of finding them.
Transcription is the process of synthesising mRNA from a DNA template. mRNA is built through polymerisation, whereby RNA polymerase adds one nucleotide to the end of the mRNA chain being built.
To initiate transcription, a transcription factor must bind to a region at the start of the gene, known as the promoter region. The promoter region contains a sequence of DNA: TATA, this is known as the TATA box. The TATA box is present in some genes and is one of the sequences that can be recognised by transcription factors. The promoter orientation determines the direction of transcription, and RNA polymerase synthesises RNA in a 5’ to 3’ direction.
Elongation begins around 35 bases downstream from the TATAAA box. One strand of DNA is used as a template strand, meaning the mRNA is complimentary to the template strand - this means regular base pairing rules apply with the exception that mRNA doesn’t use thymine (T) as a base but instead uses uracil (U) meaning A pairs with U and C pairs with G. This process continues until the entire gene is transcribed.
After transcription, mRNA is described as pre-mRNA, meaning it is not yet mature. If pre-mRNA were to be released into the cytosol, it would be quickly degraded. In order to prevent this mRNA is processed in 3 steps:
- Capping: a cap is placed on the 5’ end of the pre-mRNA strand.
- Tailing (aka polyadenylation): at the 3’ end, a tail of up to 200 adenine molecules is added to the pre-mRNA.
- Splicing: non-coding regions of pre-mRNA called introns are removed leaving only the exons which will eventually code for amino acids. Alternative splicing allows for one gene to code for multiple proteins by removing different introns depending on the final protein structure.
At this stage, the mRNA is mature and can enter the cytosol.
Translation is the process of using mature mRNA as a template for the building of a polypeptide chain. This is achieved through the use of transfer RNA (tRNA) and ribosomes and occurs in the cytoplasm.
Humans (and all other eukaryotes) have 80S ribosomes (with a 60S and 40S subunit), which are made of 4 ribosomal RNA (rRNA) molecules and 82 proteins. Prokaryotes in contrast have 70S ribosomes (with a 50S and a 30S subunit)
In order to translate mRNA to a protein, we need to convert the 4 base ‘DNA language’ to the 20 amino acid ‘protein language’ - this conversion is done through the use of tRNA.
mRNA is read at a ribosome 3 bases at a time; each set of 3 bases is called a codon. The specific set of 3 bases in the codon codes for a single specific amino acid e.g. UAC codes for tyrosine (see the diagram below).
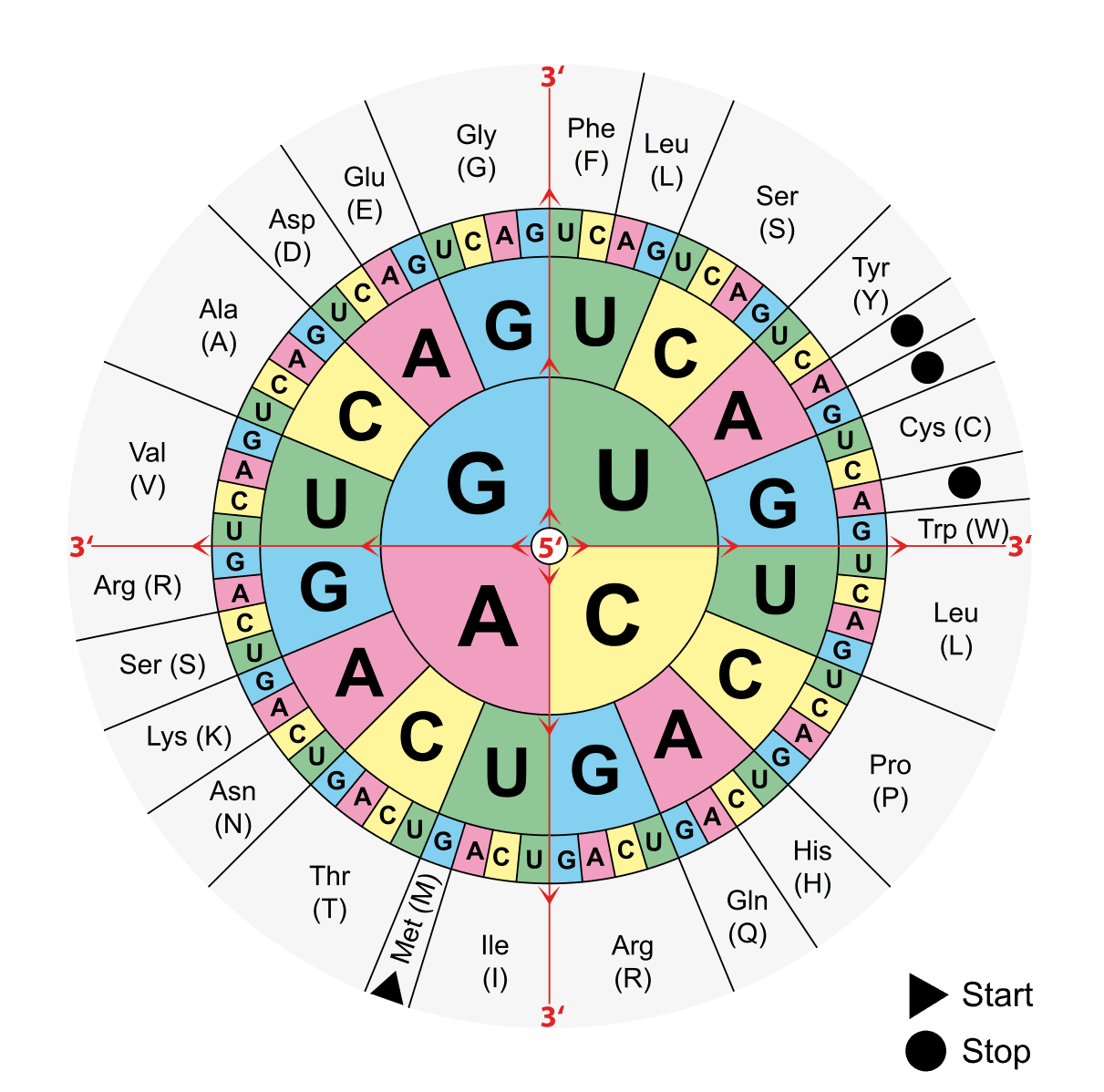
Figure: By moving from the centre of the wheel to the edge it can be determined which amino acid is coded for by a sequence of bases in a codon.
Creative commons source by Mouagip (talk) [CC BY-SA 4.0 (https://creativecommons.org/licenses/by-sa/4.0)]
As you can see from the above image, the triplet AUG on mRNA codes for methionine, which is always the first amino acid in any polypeptide chain. The following triplets on mRNA are then read, and the corresponding amino acids are added to the chain until a stop codon is reached. A stop codon is coded by either: UAA, UAG, or UGA. Stop is of course not the name of an amino acid but instead instructs the ribosome to release the finished polypeptide strand.
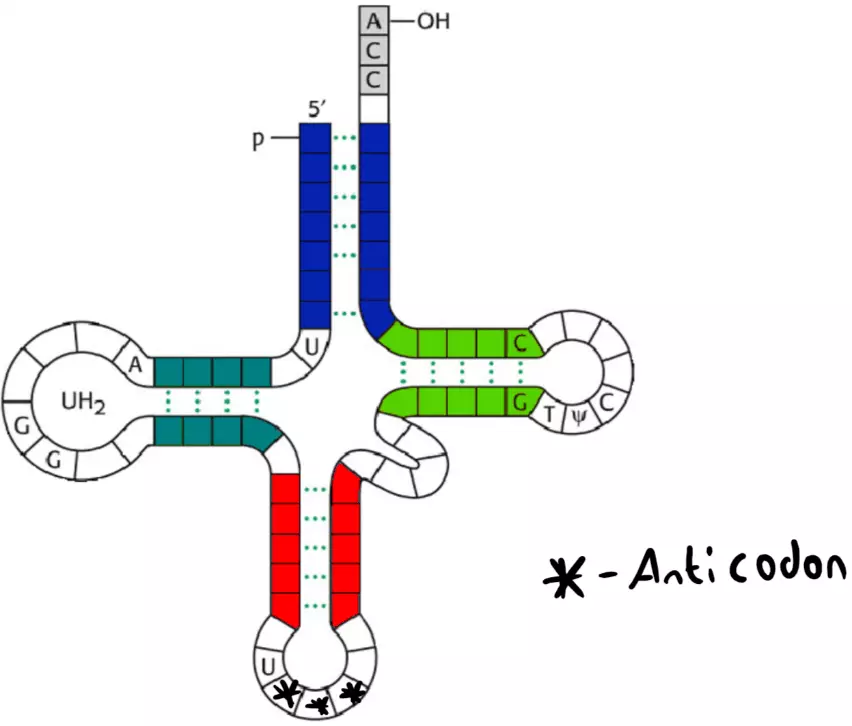
Figure: Shows the clover model of a tRNA molecule with the anticodon labelled.
Creative commons source by Yersinia~commonswiki, edited by Dr. Joshua Sturgeon [CC BY-SA 4.0 (https://creativecommons.org/licenses/by-sa/4.0)]
Above is the clover model of tRNA. The anticodon is a three bases long section on the tRNA molecule. It is called an anticodon as it is complementary to a specific codon on the mRNA strand being translated, which allows translation from the 'DNA language' to the 'protein language'. tRNA moves towards the mRNA-ribosome complex and brings with it a specific amino acid. The codon on mRNA is ‘recognised’ by the tRNA anticodon, and the attached amino acid is held in place. A peptide bond then forms between this new delivered amino acid, and the previous one in the protein chain. This process continues until a stop codon is reached.
Reviewed by: Dr. Thomas Burnell
Edited by: Dr. Ben Appleby
- 8615

