Next Lesson - Post-translational Modification and Collagen Biosynthesis
Abstract
Molecular analysis techniques can be used for the diagnosis of conditions and for a better understanding of treatment options and efficacy.
They can be split into 4 distinct types, each encompassing different techniques and analysis targets:
- Analysis of DNA at the gene level:
- Restriction enzymes (cut DNA at certain "restriction sites", allowing certain DNA sequences to be isolated).
- DNA gel electrophoresis (separate DNA fragments based on charge and size).
- Polymerase Chain Reaction (PCR) (amplify specific sequences of DNA using specific primers, allowing for further testing or to check if the primer sequence is present).
- Analysis of proteins:
- Protein gel electrophoresis (separates protein samples based on size, shape or charge).
- Immunoassays (use antibodies specific to certain antigens to test for specific proteins).
- Enzyme assays (test the function of certain enzymes by measuring their reaction rates).
- Analysis of DNA at the nucleotide level:
- DNA sequencing (determining the nucleotide order of a DNA fragment, gene or, in some cases, the entire genome, allowing analysis of genetic variation).
- Analysis of DNA at the multi-gene level:
- Southern hybridisation/Southern blotting (allows for the detection of a specific DNA sequence in a DNA sample).
- Microarrays (use thousands of fixed DNA probes on a solid surface to detect or compare gene expression or genetic variation between samples).
Core
Molecular analysis techniques are used in medicine to identify and sequence various organisms and genomes - this allows doctors to come up with diagnoses based on test results. These tests are carried out by biomedical scientists and technicians within labs and their results are key to producing accurate diagnoses that can be used for treatment.
Molecular analysis techniques can be split into 4 distinct types, each encompassing different techniques and analysis targets:
- Analysis of DNA at the gene level
- Restriction enzymes
- DNA gel electrophoresis
- Polymerase Chain Reaction (PCR)
- Analysis of proteins
- Protein gel electrophoresis
- Immunoassays
- Enzyme assays
- Analysis of DNA at the nucleotide level
- DNA sequencing
- Analysis of DNA at the multi-gene level
- Southern hybridisation/Southern blotting
- Microarrays
As a clinician, an understanding of the basics behind each technique is key in interpreting results and being able to give simple explanations to patients.
Analysis of DNA at the Gene Level
Restriction enzymes are proteins that cleave very specific DNA sequences into fragments at or near to specific recognition sites within molecules known as restriction sites - they are sometimes referred to as 'molecular scissors'. They are one type of enzyme from the broader class known as endonucleases. Bacteria produce these endonucleases as a defence mechanism against viruses, but they have been extracted and are now used commercially within diagnostic medicine.
A restriction enzyme example: Acetobacter aceti orleanensis produces a restriction enzyme, Tai I, which recognises and cuts at the restriction sites 5' ACGT / 3' TGCA:
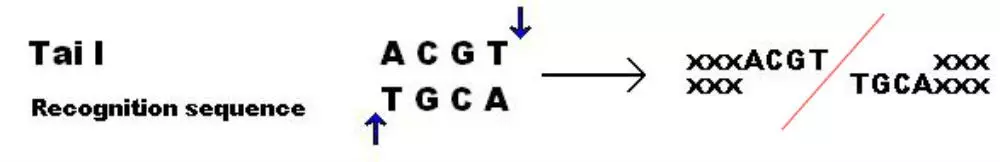
Figure - An example of a Tai I restriction enzyme cutting at its restriction sites and separating two strands of DNA. This is a commercially available restriction enzyme used in assays
Creative commons source by Inks002 at the English Wikipedia [CC BY-SA 3.0 (http://creativecommons.org/licenses/by-sa/3.0/)]
Isolated restriction enzymes are used to manipulate DNA for different medical applications, for example:
- DNA/Gene Cloning
- Restriction enzymes can be used to assist in the insertion of specific genes into plasmid (small circular DNA segments found in bacteria) vectors during gene cloning and protein production experiments (eg. inserting the human insulin gene into an E. coli plasmid, allowing it to produce human insulin).
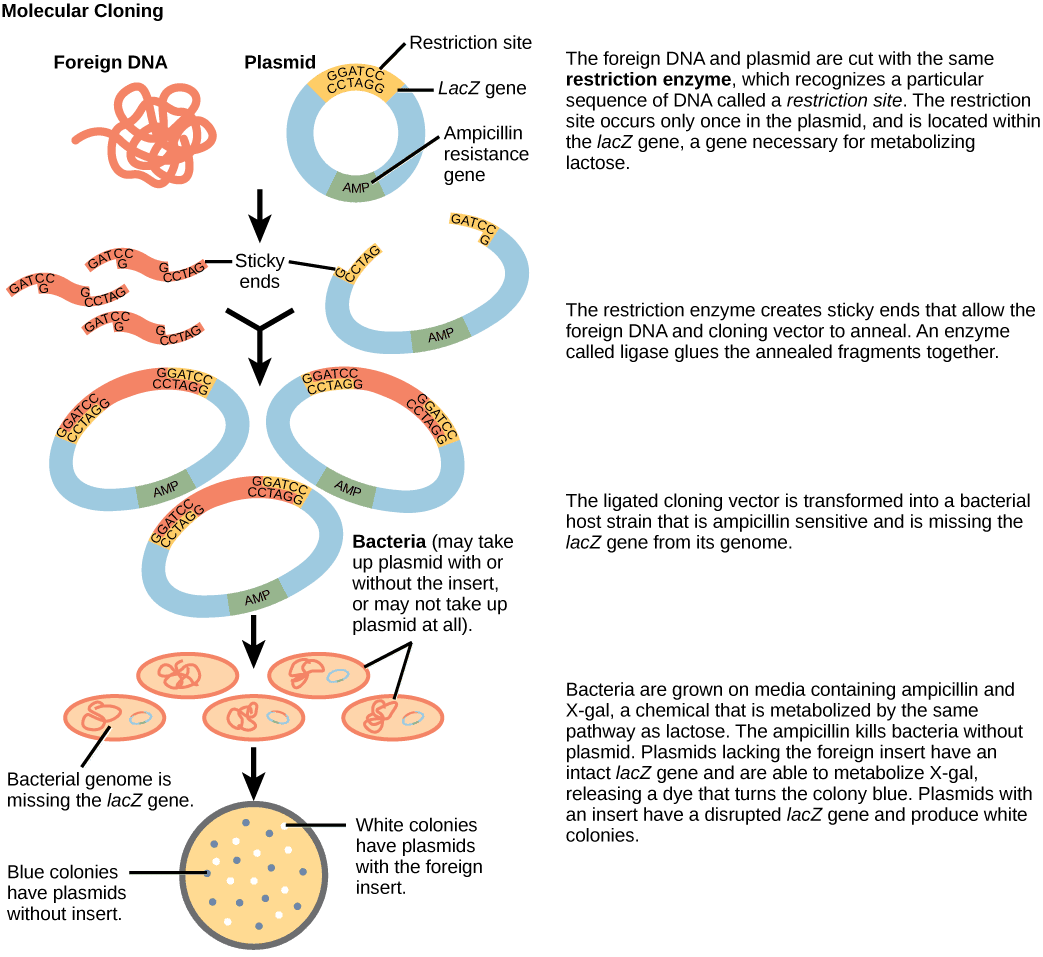
Diagram - A step-by-step example of how a restriction enzyme is used to add foreign DNA (eg. the human insulin gene) to a bacterial plasmid
Creative commons source by CNX OpenStax [CC BY 4.0 (https://creativecommons.org/licenses/by/4.0)]
- Diagnosis of single base changes
- The main diagnostic use of restriction enzymes is in being used to distinguish gene alleles by specifically recognising single base changes in DNA known as single nucleotide polymorphisms (SNPs), when combined with gel electrophoresis separation. The different lengths of DNA generated by restriction digest produce a specific pattern of bands after gel electrophoresis, and can also be used for DNA fingerprinting.
- Southern Blotting (covered later)
- In a similar manner to above, restriction enzymes are used to digest genomic DNA for gene analysis by Southern blot. This technique allows researchers to identify how many copies of a gene are present in the genome of one individual, or how many gene mutations (polymorphisms) have occurred within a population.
DNA gel electrophoresis is a technique used to separate DNA fragments according to their size.
A DNA gel electrophoresis setup contains 4 main components:
- Gel (eg. agarose/ polyacrylamide) - A matrix that allows the separation of DNA fragments.
- Buffer (eg. TAE or TBE, which contain EDTA) - Provides ions to allow an electric current to pass across the gel and helps maintain a stable pH.
- Power supply - Generates an electrical charge difference across the gel.
- Stain/detection (eg. Ethidium bromide or other DNA-binding dyes) - To easily identify the presence of the separated DNA.
All DNA is negatively charged and will move towards the anode if placed in a gel electrophoresis electric field. Because all DNA fragments have the same amount of charge per mass, small fragments move through the gel faster than large ones; therefore, DNA fragments can be separated on the basis of size.
DNA gel electrophoresis is used with restriction enzymes to identify certain DNA sequences within genes.
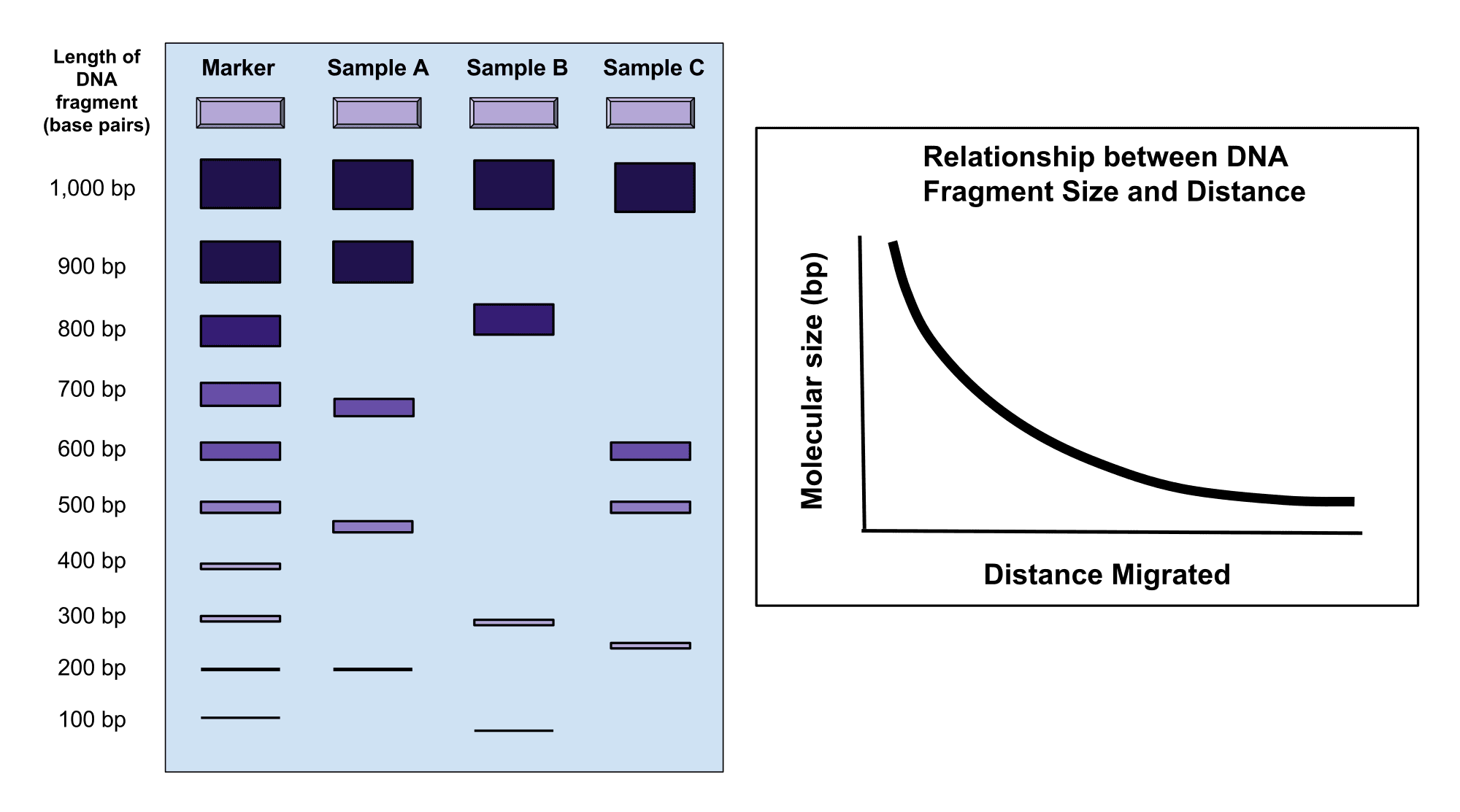
Figure - Demonstrates what a DNA gel electrophoresis would look like - samples are compared based on size, with the distance they travel down the sheet based on how large they are
Creative commons source by Mckenzielower [CC BY-SA 4.0 (https://creativecommons.org/licenses/by-sa/4.0)]
Polymerase Chain Reaction (PCR)
PCR is a very common procedure used in the amplification (making more of) a target sequence of DNA (or RNA in RT-PCR). This is carried out using a special thermostable DNA polymerase (Taq Pol), able to operate at temperatures above what a normal polymerase would function at (normally enzymes denature at high temperatures).
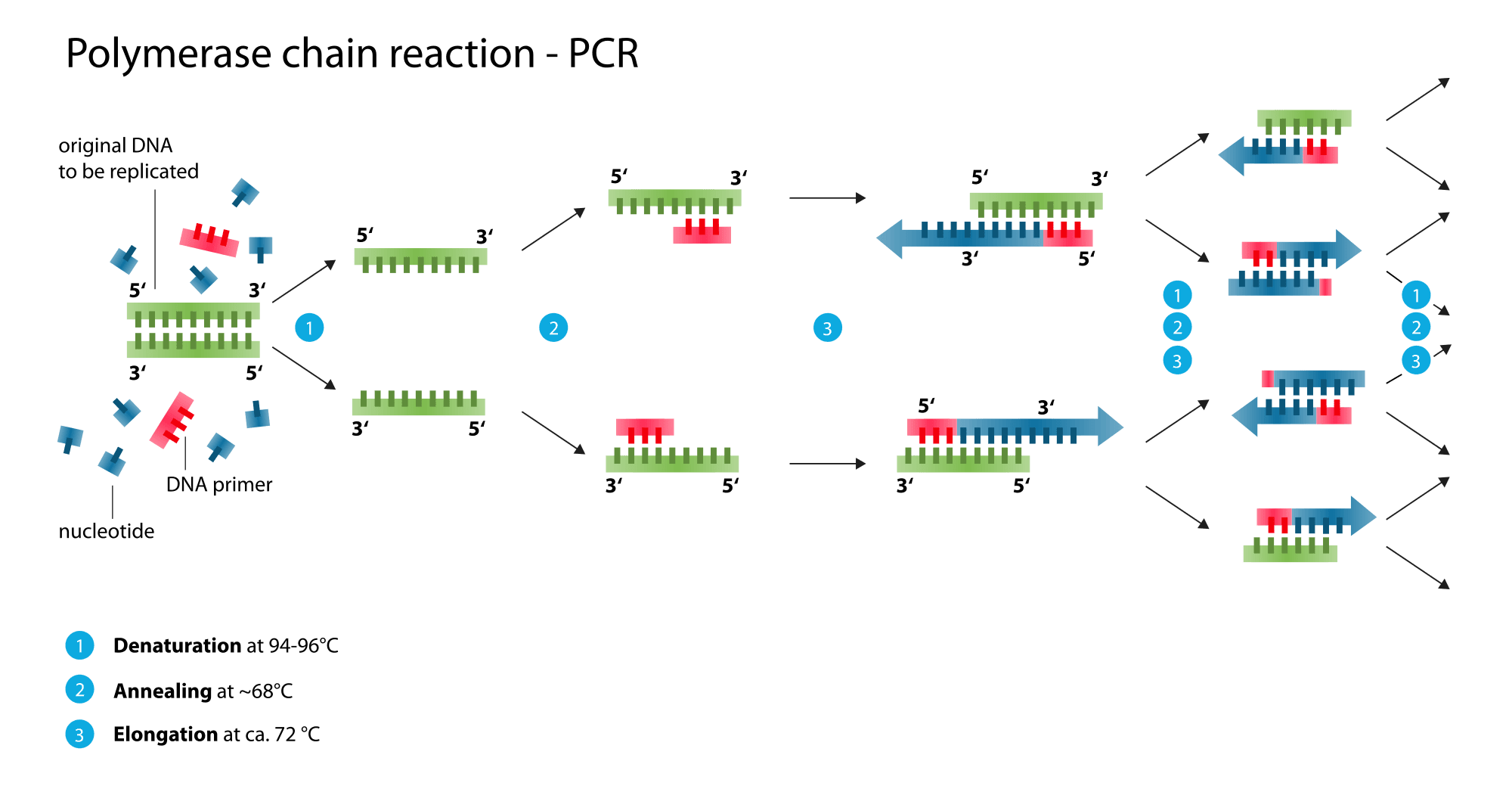
Diagram - The step-by-step sequence for amplifying DNA using PCR - this is repeated in cycles until sufficient DNA has been copied
Creative commons source by Enzoklop [CC BY-SA 3.0 (https://creativecommons.org/licenses/by-sa/3.0)]
A pair of primers (forward and reverse) are created, uniquely defining the sequence of DNA to be copied from the DNA sample being tested. A cycle is then setup, whereby the temperature is raised, causing DNA denaturing (1), then the temperature is reduced, allowing re-annealing (2) and polymerisation/elongation (3). This repeated cycle results in exponential increase in DNA.
PCR has many medical uses:
- To amplify a specific DNA fragment
- To investigate single base mutations e.g. Tay Sachs, Sickle Cell disease
- To investigate small deletions or insertions e.g. Cystic Fibrosis
- To investigate variation, genetic relationships e.g. DNA profiling, DNA typing
Protein gel electrophoresis is a technique used to separate protein fragments according to their size, shape or charge.
Similarly to a DNA gel electrophoresis, a protein gel electrophoresis setup contains 4 main components:
- Gel (eg. agarose/ polyacrylamide) - A matrix that allows separation of the protein sample.
- Buffer (eg. Tris-glycine buffer, which may contain EDTA) - Provides ions for current flow and helps maintain pH during electrophoresis.
- Power supply - Generates charge difference across the gel.
- Stain/detection (eg. Coomassie Blue) - To identify the presence of the separated proteins.
Proteins are charged molecules and will move towards the anode or the cathode if placed in an electric field. This charge is not equal in all proteins and therefore proteins can be separated on the basis of size, shape or charge.
Serum proteins from a patient can be taken and run through a protein gel electrophoresis to identify certain pathogenic markers, such as certain enzymes or antibodies.
There are 3 main variations of Protein Gel Electrophoresis, with each separating based on different protein properties:
- Sodium dodecyl sulphate polyacrylamide gel electrophoresis (SDS-PAGE)
- Separates proteins based on size.
- Isoelectric focusing
- Separates proteins on the basis of charge - proteins will migrate up a pH gradient until they reach a pH equal to their pI (isoelectric point - the pH at which a molecule carries no net electrical charge), where there will be no net charge and so the proteins will stop migrating.
- Two-dimensional electrophoresis (2D-PAGE)
- Allows the separation of complex mixtures of proteins and is particularly important for diagnosing disease states in different tissues.
Immunoassays rely on the properties of antibodies. Antibodies bind to specific protein targets (known as antigens) - they only recognise and bind to a few amino acids in a sequence on the protein (the epitope of the antigen) and so can be very sensitive to small changes in protein structure.
Antibodies can be one of two types:
- Polyclonal antibodies
- Produced by many different B lymphocytes and are all slightly different, but are all specific to 1 antigen, where they are able to recognise multiple epitopes on that antigen.
- Monoclonal antibodies
- Produced from a single B lymphocyte and are all identical - they are specific to 1 antigen and only 1 epitope on that antigen.
Immunoassay examples:
- Western Blotting (aka protein immunoblotting)
- A protein sample is denatured and run through gel electrophoresis.
- A "primary" antibody is added that binds to a specific target protein (if the specific protein that the investigator is looking for is present in the sample).
- The electrophoresis membrane is incubated in a solution containing the primary antibody, after which excess antibody is washed off.
- A "secondary" antibody is added which recognises and binds to the primary antibody (if it found the specific protein being investigated and bound).
- The secondary antibody is able to fluoresce or is stained and so can be visually seen if it attaches to the protein.
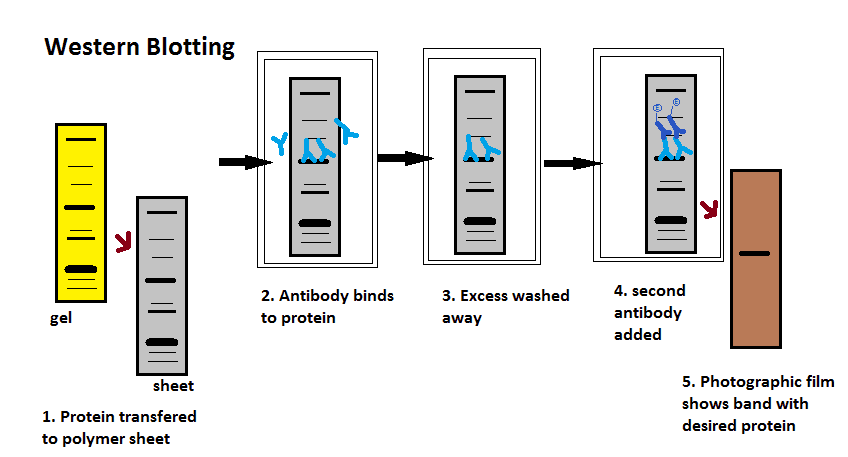
Diagram - The steps of performing a Western Blot with the expected outcome
Creative commons source by Cawang [CC BY-SA 4.0 (https://creativecommons.org/licenses/by-sa/4.0)]
- Enzyme-linked immunoabsorbent assay (ELISA)
- An antibody with specificity for a particular antigen is chosen.
- The sample with an unknown amount of antigen is immobilised on a microtitre plate.
- After the antigen is added, the detection antibody is added, forming a complex with the antigen.
- This "primary" or detection antibody can be covalently linked to an enzyme or can itself be detected by a secondary antibody that is linked to an enzyme.
- This is repeated several times, after which an enzymatic substrate is added to produce a visible signal, which indicates the quantity of antigen in the sample.
- A good use example is measuring cardiac troponin I (cTnI) levels in the blood after a suspected acute myocardial infarction, which contributes to the diagnosis of MI when interpreted alongside the clinical picture and ECG findings.
Enzyme assays are used for measuring enzymatic activity, particularly enzyme kinetics and enzyme inhibition. They generally involve measuring the product of an enzymatic reaction and the rate at which this is formed.
They can be split into 2 types:
- Continuous assays - the most convenient, requiring only one assay to give a result.
- Spectrophotometry - measurement of the change in the amount of light the solution absorbs.
- Chemoluminescence - measurement of the amount of light emitted by the reaction.
- Discontinuous assays - samples are taken from the reaction and measured at various times - more effort, but can be more sensitive.
- Radioactivity.
- Chromatography.
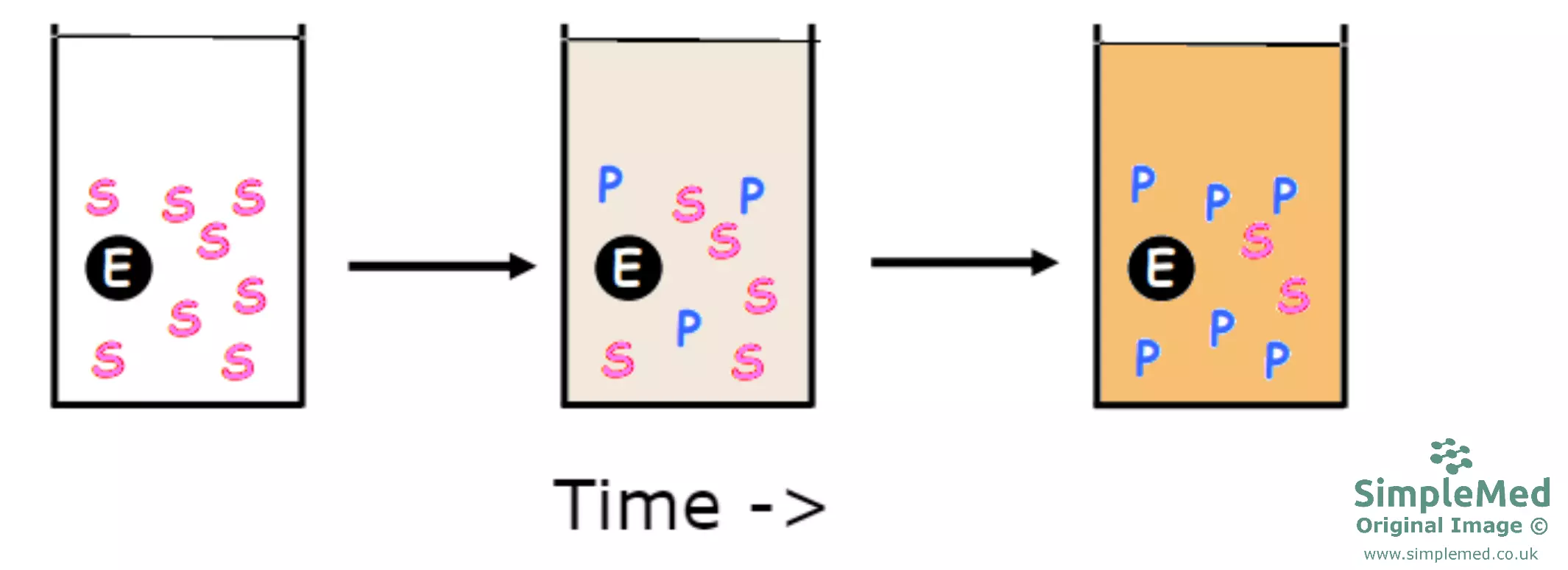
Diagram - Example of how an enzyme assay occurs. The substrate is converted to product with the help of the enzyme. In this case, a spectrophotometric enzyme assay is being carried out, measuring the opacity of the solution created. As more product is formed, the solution becomes more opaque, a measurement of the reaction rate and thus kinetics of the enzyme
SimpleMed original by Dr. Marcus Judge
Important use examples of certain serum enzyme measurements include:
- Aspartate transaminase (AST)/ Alanine transaminase (ALT) - Markers for liver damage/ disease
- Amylase/ lipase - Marker for pancreatitis.
- gamma-glutamyl transferase (GGT) - Marker for liver damage and biliary tract obstruction, also increased by alcohol.
- Alkaline phosphatase (ALP) - Marker for bone disorders and bile tract obstruction.
Analysis of DNA at the Nucleotide Level
DNA sequencing is the process of determining the nucleic acid sequence of a genome - the order of nucleotides within DNA. It allows the identification and visualisation of entire genomes and is evolving quickly into a diagnostic tool, with reduced costs and increased speeds (Next-Gen sequencing).
There are many different methods of carrying out this process that are out of the scope of a standard medical professional's required knowledge, but an understanding of the ethics of identifying genetic variants across a person's genome is an active area of debate.
Analysis of DNA at the Multi-Gene Level
A Southern blot is an analysis method used in molecular biology for the detection of a specific DNA sequence in DNA samples. Southern blotting combines the transfer of electrophoresis-separated DNA fragments to a (nitrocellulose) filter membrane and then the subsequent fragment detection by probe hybridisation (a single DNA fragment with a target-allele specific sequence whose presence in the target DNA is being determined). The pattern of hybridisation is visualised on X-ray film by autoradiography (in the case of a radioactive or fluorescent probe), or by development of colour on the membrane (if a fluorescence detection method is used).
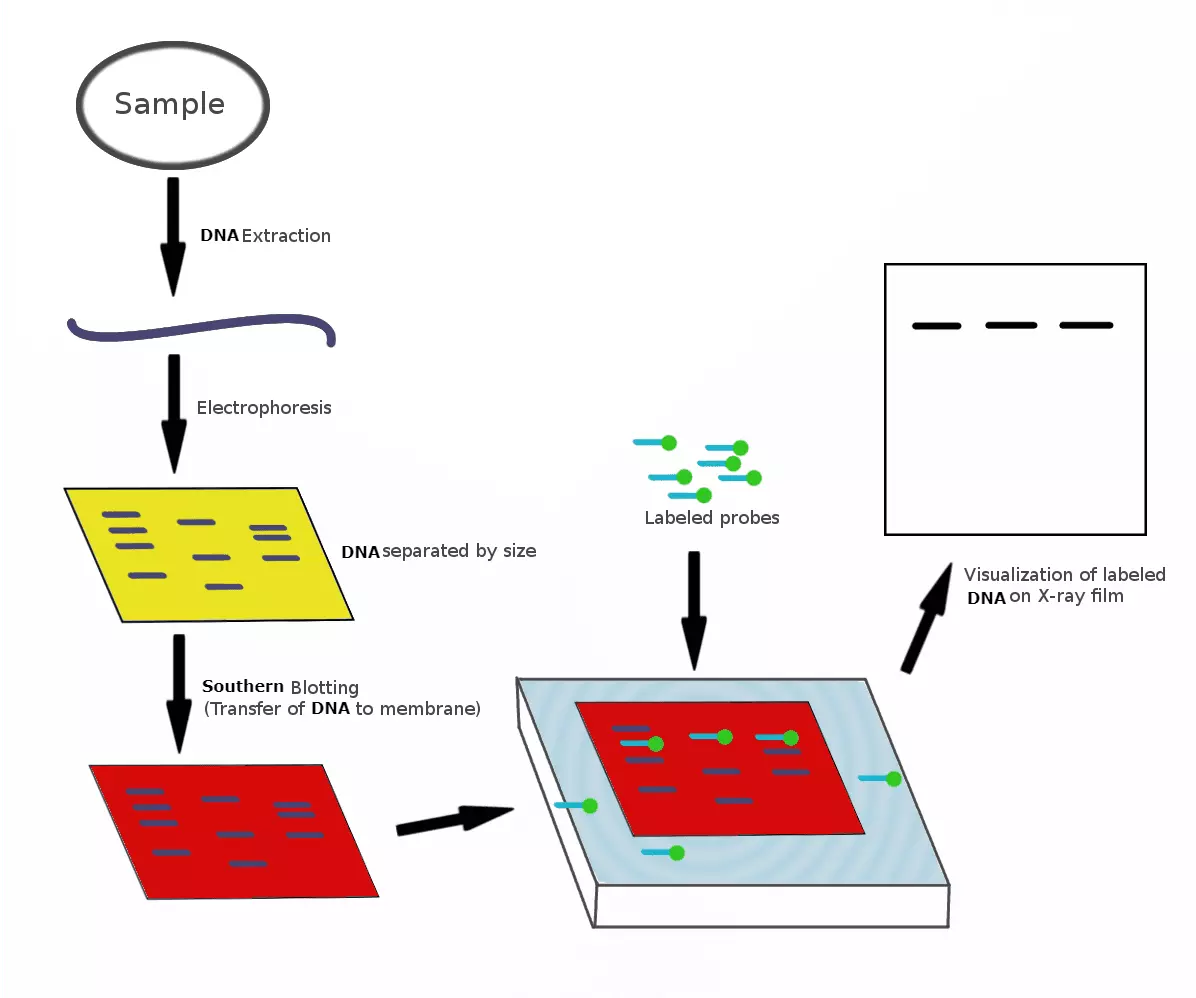
Diagram - Step-by-step showing how a Southern Blot is carried out to visualise if the DNA tested for is present in the sample
Creative commons source by Ilewieszoośmiornicach, edited by Dr. Marcus Judge [CC BY-SA 4.0 (https://creativecommons.org/licenses/by-sa/4.0)]
A DNA microarray is a collection of microscopic DNA spots attached to a solid surface. Microarrays are a modern technique used to analyse thousands of predefined DNA sequences from a single sample on one solid surface. They use fluorescent probe hybridisation (similar to a Southern blot), and differences in gene presence or expression between samples are identified by comparing fluorescence signal intensity using dedicated analysis software.
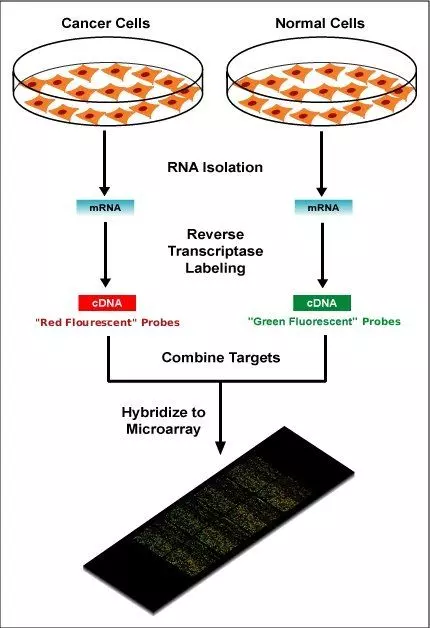

Diagram - How a microarray is created. Below it, a comparison of 4 separate DNA microarrays from 4 different genomes - notice how it is possible to compare between them all for the presence of certain DNA probes and thus certain DNA sequences
Public Domain Images
DNA fingerprinting is a forensic technique in criminal investigations (eg. comparing criminal suspects' profiles to DNA evidence so as to assess the likelihood of their involvement in the crime), parentage testing and in genealogical (can show family relationships) and medical research. It uses small samples such as hair or a mouth swab, from which DNA is extracted, amplified (for example by PCR), and then restricted and analysed for specific patterns.
Karyotyping is the process by which metaphase chromosomes are photographed and looked at under a light microscope in order to determine the chromosomes present within an individual's genome, as well as their chromosome shape and number.
Attention is paid to their length, the position of the centromeres, banding pattern, any differences between the sex chromosomes, and any other physical characteristics. The preparation and study of karyotypes is part of cytogenetics.
The basic number of chromosomes in the somatic cells of a species is called the somatic number and is designated 2n. In the germ-line (the sex cells) the chromosome number is n (humans: n = 23 - the sex cells must merge with another sex cell to form a full complement of chromosomes). Therefore, in humans 2n = 46. A normal human will have 46 chromosomes (2 of which are sex chromosomes), which is represented as such:
Males: 46, XY
Females: 46, XX
A patient with Down's Syndrome will have Trisomy 21 (three copies of chromosome 21) and thus their karyotype number will look like:
Males: 47, XY, +21
Females: 47, XX, +21

Image - A normal human male karyotype, showing 46 chromosomes
Public Domain Image
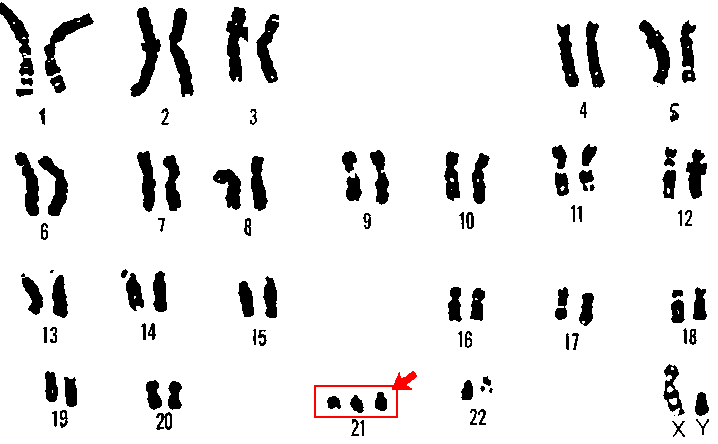
Image - The karyotype of a male with trisomy 21 (Down's Syndrome) - notice the 3 chromosome 21s present
Public Domain Image
Fluorescent In Situ Hybridisation (FISH)
Fluorescence in situ hybridization (FISH) is a molecular cytogenetic technique that uses fluorescent probes that bind to only those parts of a nucleic acid sequence with a high degree of sequence complementarity. Fluorescence microscopy is then used to find out where the fluorescent probe is bound to the chromosomes. FISH can also be used to detect and localise specific RNA targets in cells, circulating tumour cells, and tissue samples.
FISH can be used to form a diagnosis, to evaluate prognosis, or to evaluate remission of a disease, such as cancer. Treatment can then be specifically tailored. FISH can also be used to detect diseased cells more easily than standard cytogenetic methods, which require dividing cells and labour and time-intensive manual preparation and analysis of the slides by a technologist.
- 11112

